Chemical Decorations of "MARs" Residents in Orchestrating Eukaryotic Gene Regulation
- PMID: 33409278
- PMCID: PMC7779526
- DOI: 10.3389/fcell.2020.602994
Chemical Decorations of "MARs" Residents in Orchestrating Eukaryotic Gene Regulation
Abstract
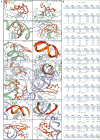
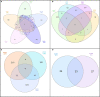
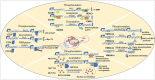
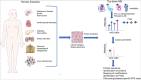
Genome organization plays a crucial role in gene regulation, orchestrating multiple cellular functions. A meshwork of proteins constituting a three-dimensional (3D) matrix helps in maintaining the genomic architecture. Sequences of DNA that are involved in tethering the chromatin to the matrix are called scaffold/matrix attachment regions (S/MARs), and the proteins that bind to these sequences and mediate tethering are termed S/MAR-binding proteins (S/MARBPs). The regulation of S/MARBPs is important for cellular functions and is altered under different conditions. Limited information is available presently to understand the structure-function relationship conclusively. Although all S/MARBPs bind to DNA, their context- and tissue-specific regulatory roles cannot be justified solely based on the available information on their structures. Conformational changes in a protein lead to changes in protein-protein interactions (PPIs) that essentially would regulate functional outcomes. A well-studied form of protein regulation is post-translational modification (PTM). It involves disulfide bond formation, cleavage of precursor proteins, and addition or removal of low-molecular-weight groups, leading to modifications like phosphorylation, methylation, SUMOylation, acetylation, PARylation, and ubiquitination. These chemical modifications lead to varied functional outcomes by mechanisms like modifying DNA-protein interactions and PPIs, altering protein function, stability, and crosstalk with other PTMs regulating subcellular localizations. S/MARBPs are reported to be regulated by PTMs, thereby contributing to gene regulation. In this review, we discuss the current understanding, scope, disease implications, and future perspectives of the diverse PTMs regulating functions of S/MARBPs.
Keywords: S/MAR-binding protein; chromatin 3D architecture; disease; gene regulation; post-translation modification (PTM).
Copyright © 2020 Roychowdhury and Chattopadhyay.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

