Complex Evolution of the Mismatch Repair System in Eukaryotes is Illuminated by Novel Archaeal Genomes
- PMID: 33409543
- PMCID: PMC7884376
- DOI: 10.1007/s00239-020-09979-5
Complex Evolution of the Mismatch Repair System in Eukaryotes is Illuminated by Novel Archaeal Genomes
Abstract
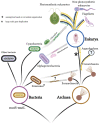
Repairing DNA damage is one of the most important functions of the 'housekeeping' proteins, as DNA molecules are constantly subject to different kinds of damage. An important mechanism of DNA repair is the mismatch repair system (MMR). In eukaryotes, it is more complex than it is in bacteria or Archaea due to an inflated number of paralogues produced as a result of an extensive process of gene duplication and further specialization upon the evolution of the first eukaryotes, including an important part of the meiotic machinery. Recently, the discovery and sequencing of Asgard Archaea allowed us to revisit the MMR system evolution with the addition of new data from a group that is closely related to the eukaryotic ancestor. This new analysis provided evidence for a complex evolutionary history of eukaryotic MMR: an archaeal origin for the nuclear MMR system in eukaryotes, with subsequent acquisitions of other MMR systems from organelles.
Keywords: Asgard Archaea; DNA repair; Eukaryotes; Mismatch repair; mutL; mutS.
Figures


Similar articles
-
Asgard archaea illuminate the origin of eukaryotic cellular complexity.Nature. 2017 Jan 19;541(7637):353-358. doi: 10.1038/nature21031. Epub 2017 Jan 11. Nature. 2017. PMID: 28077874
-
Origin of eukaryotes from within archaea, archaeal eukaryome and bursts of gene gain: eukaryogenesis just made easier?Philos Trans R Soc Lond B Biol Sci. 2015 Sep 26;370(1678):20140333. doi: 10.1098/rstb.2014.0333. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26323764 Free PMC article. Review.
-
Expanded diversity of Asgard archaea and their relationships with eukaryotes.Nature. 2021 May;593(7860):553-557. doi: 10.1038/s41586-021-03494-3. Epub 2021 Apr 28. Nature. 2021. PMID: 33911286 Free PMC article.
-
The ambiguity of the basic terms related to eukaryotes and the more consistent etymology based on eukaryotic signatures in Asgard archaea.Biosystems. 2020 Nov;197:104178. doi: 10.1016/j.biosystems.2020.104178. Epub 2020 Jun 10. Biosystems. 2020. PMID: 32534168
-
Archaea and the origin of eukaryotes.Nat Rev Microbiol. 2017 Nov 10;15(12):711-723. doi: 10.1038/nrmicro.2017.133. Nat Rev Microbiol. 2017. PMID: 29123225 Review.
Cited by
-
Matreex: Compact and Interactive Visualization for Scalable Studies of Large Gene Families.Genome Biol Evol. 2024 Jun 4;16(6):evae100. doi: 10.1093/gbe/evae100. Genome Biol Evol. 2024. PMID: 38742690 Free PMC article.
-
Intermolecular Gene Conversion for the Equalization of Genome Copies in the Polyploid Haloarchaeon Haloferax volcanii: Identification of Important Proteins.Genes (Basel). 2024 Jul 1;15(7):861. doi: 10.3390/genes15070861. Genes (Basel). 2024. PMID: 39062640 Free PMC article.
-
Expansion of the MutS gene family in plants.Plant Cell. 2025 Jul 1;37(7):koae277. doi: 10.1093/plcell/koae277. Plant Cell. 2025. PMID: 39692564 Free PMC article.
-
Expansion of the MutS Gene Family in Plants.bioRxiv [Preprint]. 2024 Jul 20:2024.07.17.603841. doi: 10.1101/2024.07.17.603841. bioRxiv. 2024. Update in: Plant Cell. 2025 Jul 1;37(7):koae277. doi: 10.1093/plcell/koae277. PMID: 39071318 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

