In silico comparative analysis of Aeromonas Type VI Secretion System
- PMID: 33410103
- PMCID: PMC7966616
- DOI: 10.1007/s42770-020-00405-y
In silico comparative analysis of Aeromonas Type VI Secretion System
Abstract
Aeromonas are bacteria broadly spread in the environment, particularly in aquatic habitats and can induce human infections. Several virulence factors have been described associated with bacterial pathogenicity, such as the Type VI Secretion System (T6SS). This system translocates effector proteins into target cells through a bacteriophage-like contractile structure encoded by tss genes. Here, a total of 446 Aeromonas genome sequences were screened for T6SS and the proteins subjected to in silico analysis. The T6SS-encoding locus was detected in 243 genomes and its genes are encoded in a cluster containing 13 core and 5 accessory genes, in highly conserved synteny. The amino acid residues identity of T6SS proteins ranges from 78 to 98.8%. In most strains, a pair of tssD and tssI is located upstream the cluster (tssD-2, tssI-2) and another pair was detected distant from the cluster (tssD-1, tssI-1). Significant variability was seen in TssI (VgrG) C-terminal region, which was sorted in four groups based on its sequence length and protein domains. TssI containing ADP-ribosyltransferase domain are associated exclusively with TssI-1, while genes coding proteins carrying DUF4123 (a conserved domain of unknown function) were observed downstream tssI-1 or tssI-2 and escort of possible effector proteins. Genes coding proteins containing DUF1910 and DUF1911 domains were located only downstream tssI-2 and might represent a pair of toxin/immunity proteins. Nearly all strains display downstream tssI-3, that codes for a lysozyme family domain protein. These data reveal that Aeromonas T6SS cluster synteny is conserved and the low identity observed for some genes might be due to species heterogeneity or its niche/functionality.
Keywords: Aeromonas; Comparative genomic analysis; Type VI Secretion System; tss genes.
Figures




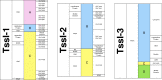

 type A,
type A,  type B,
type B,  type C, and
type C, and  type D; protein-coding genes upstream and downstream are also indicated, relative to tss cluster or tssDI copies. The number sign indicates gene coding for PAAR domain–containing protein
type D; protein-coding genes upstream and downstream are also indicated, relative to tss cluster or tssDI copies. The number sign indicates gene coding for PAAR domain–containing proteinReferences
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

