A nidovirus perspective on SARS-CoV-2
- PMID: 33413979
- PMCID: PMC7664520
- DOI: 10.1016/j.bbrc.2020.11.015
A nidovirus perspective on SARS-CoV-2
Abstract
Two pandemics of respiratory distress diseases associated with zoonotic introductions of the species Severe acute respiratory syndrome-related coronavirus in the human population during 21st century raised unprecedented interest in coronavirus research and assigned it unseen urgency. The two viruses responsible for the outbreaks, SARS-CoV and SARS-CoV-2, respectively, are in the spotlight, and SARS-CoV-2 is the focus of the current fast-paced research. Its foundation was laid down by studies of many corona- and related viruses that collectively form the vast order Nidovirales. Comparative genomics of nidoviruses played a key role in this advancement over more than 30 years. It facilitated the transfer of knowledge from characterized to newly identified viruses, including SARS-CoV and SARS-CoV-2, as well as contributed to the dissection of the nidovirus proteome and identification of patterns of variations between different taxonomic groups, from species to families. This review revisits selected cases of protein conservation and variation that define nidoviruses, illustrates the remarkable plasticity of the proteome during nidovirus adaptation, and asks questions at the interface of the proteome and processes that are vital for nidovirus reproduction and could inform the ongoing research of SARS-CoV-2.
Keywords: Comparative genomics; Coronaviruses; Evolution; Nidoviruses; Proteome.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
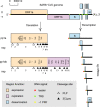
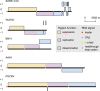


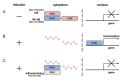
Similar articles
-
The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2.Nat Microbiol. 2020 Apr;5(4):536-544. doi: 10.1038/s41564-020-0695-z. Epub 2020 Mar 2. Nat Microbiol. 2020. PMID: 32123347 Free PMC article.
-
Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes.PLoS Pathog. 2011 Sep;7(9):e1002215. doi: 10.1371/journal.ppat.1002215. Epub 2011 Sep 8. PLoS Pathog. 2011. PMID: 21931546 Free PMC article.
-
Insight into the evolution of nidovirus endoribonuclease based on the finding that nsp15 from porcine Deltacoronavirus functions as a dimer.J Biol Chem. 2018 Aug 3;293(31):12054-12067. doi: 10.1074/jbc.RA118.003756. Epub 2018 Jun 10. J Biol Chem. 2018. PMID: 29887523 Free PMC article.
-
The relationship of severe acute respiratory syndrome coronavirus with avian and other coronaviruses.Avian Dis. 2006 Sep;50(3):315-20. doi: 10.1637/7612-042006R.1. Avian Dis. 2006. PMID: 17039827 Review.
-
Molecular Basis of Pathogenesis of Coronaviruses: A Comparative Genomics Approach to Planetary Health to Prevent Zoonotic Outbreaks in the 21st Century.OMICS. 2020 Nov;24(11):634-644. doi: 10.1089/omi.2020.0131. Epub 2020 Sep 16. OMICS. 2020. PMID: 32940573 Review.
Cited by
-
Molecular Dynamics Simulations to Decipher the Role of Phosphorylation of SARS-CoV-2 Nonstructural Proteins (nsps) in Viral Replication.Viruses. 2022 Nov 2;14(11):2436. doi: 10.3390/v14112436. Viruses. 2022. PMID: 36366534 Free PMC article.
-
Viral and cellular translation during SARS-CoV-2 infection.FEBS Open Bio. 2022 Sep;12(9):1584-1601. doi: 10.1002/2211-5463.13413. Epub 2022 Apr 25. FEBS Open Bio. 2022. PMID: 35429230 Free PMC article. Review.
-
Impact of SARS-CoV-2 Mutations on PCR Assay Sequence Alignment.Front Public Health. 2022 Apr 28;10:889973. doi: 10.3389/fpubh.2022.889973. eCollection 2022. Front Public Health. 2022. PMID: 35570946 Free PMC article.
-
Duplex One-Step RT-qPCR Assays for Simultaneous Detection of Genomic and Subgenomic RNAs of SARS-CoV-2 Variants.Viruses. 2022 May 17;14(5):1066. doi: 10.3390/v14051066. Viruses. 2022. PMID: 35632807 Free PMC article.
-
Coronavirus 2'-O-methyltransferase: A promising therapeutic target.Virus Res. 2023 Oct 15;336:199211. doi: 10.1016/j.virusres.2023.199211. Epub 2023 Sep 4. Virus Res. 2023. PMID: 37634741 Free PMC article. Review.
References
-
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., for China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Li M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T.K., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. - DOI - PMC - PubMed
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G., Shi Z.-L. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. - DOI - PMC - PubMed
-
- S.G. Siddell (Ed.), The Coronaviridae, in: The Viruses, H. Fraenkel-Conrat, R.R. Wagner (Eds.), Plenum Press, New York, 1995, pp. XVIII-418.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

