KDM1A maintains genome-wide homeostasis of transcriptional enhancers
- PMID: 33414108
- PMCID: PMC7849409
- DOI: 10.1101/gr.234559.118
KDM1A maintains genome-wide homeostasis of transcriptional enhancers
Abstract
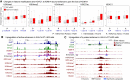
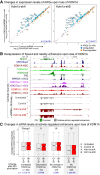
Transcriptional enhancers enable exquisite spatiotemporal control of gene expression in metazoans. Enrichment of monomethylation of histone H3 lysine 4 (H3K4me1) is a major chromatin signature of transcriptional enhancers. Lysine (K)-specific demethylase 1A (KDM1A, also known as LSD1), an H3K4me2/me1 demethylase, inactivates stem-cell enhancers during the differentiation of mouse embryonic stem cells (mESCs). However, its role in undifferentiated mESCs remains obscure. Here, we show that KDM1A actively maintains the optimal enhancer status in both undifferentiated and lineage-committed cells. KDM1A occupies a majority of enhancers in undifferentiated mESCs. KDM1A levels at enhancers exhibit clear positive correlations with its substrate H3K4me2, H3K27ac, and transcription at enhancers. In Kdm1a-deficient mESCs, a large fraction of these enhancers gains additional H3K4 methylation, which is accompanied by increases in H3K27 acetylation and increased expression of both enhancer RNAs (eRNAs) and target genes. In postmitotic neurons, loss of KDM1A leads to premature activation of neuronal activity-dependent enhancers and genes. Taken together, these results suggest that KDM1A is a versatile regulator of enhancers and acts as a rheostat to maintain optimal enhancer activity by counterbalancing H3K4 methylation at enhancers.
© 2021 Agarwal et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





References
-
- Acampora D, Omodei D, Petrosino G, Garofalo A, Savarese M, Nigro V, Di Giovannantonio LG, Mercadante V, Simeone A. 2016. Loss of the Otx2-binding site in the Nanog promoter affects the integrity of embryonic stem cell subtypes and specification of inner cell mass-derived epiblast. Cell Rep 15: 2651–2664. 10.1016/j.celrep.2016.05.041 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
