Characterization of a common progenitor pool of the epicardium and myocardium
- PMID: 33414188
- PMCID: PMC7615359
- DOI: 10.1126/science.abb2986
Characterization of a common progenitor pool of the epicardium and myocardium
Abstract
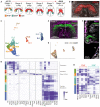
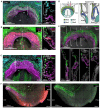
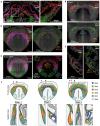
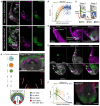
The mammalian heart is derived from multiple cell lineages; however, our understanding of when and how the diverse cardiac cell types arise is limited. We mapped the origin of the embryonic mouse heart at single-cell resolution using a combination of transcriptomic, imaging, and genetic lineage labeling approaches. This mapping provided a transcriptional and anatomic definition of cardiac progenitor types. Furthermore, it revealed a cardiac progenitor pool that is anatomically and transcriptionally distinct from currently known cardiac progenitors. Besides contributing to cardiomyocytes, these cells also represent the earliest progenitor of the epicardium, a source of trophic factors and cells during cardiac development and injury. This study provides detailed insights into the formation of early cardiac cell types, with particular relevance to the development of cell-based cardiac regenerative therapies.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures

References
-
- Lawson Ka. Fate mapping the mouse embryo. Int J Dev Biol. 1999;43:773–775. - PubMed
-
- Parameswaran M, Tam PPL. Regionalisation of cell fate and morphogenetic movement of the mesoderm during mouse gastrulation. Dev Genet. 1995;17:16–28. - PubMed
-
- Saga Y, et al. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. - PubMed
-
- Meilhac SM, Buckingham ME. The deployment of cell lineages that form the mammalian heart. Nat Rev Cardiol. 2018;15:705–724. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

