Structural basis for antibody inhibition of flavivirus NS1-triggered endothelial dysfunction
- PMID: 33414220
- PMCID: PMC8000976
- DOI: 10.1126/science.abc0476
Structural basis for antibody inhibition of flavivirus NS1-triggered endothelial dysfunction
Abstract
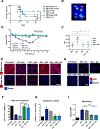
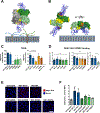
Medically important flaviviruses cause diverse disease pathologies and collectively are responsible for a major global disease burden. A contributing factor to pathogenesis is secreted flavivirus nonstructural protein 1 (NS1). Despite demonstrated protection by NS1-specific antibodies against lethal flavivirus challenge, the structural and mechanistic basis remains unknown. Here, we present three crystal structures of full-length dengue virus NS1 complexed with a flavivirus-cross-reactive, NS1-specific monoclonal antibody, 2B7, at resolutions between 2.89 and 3.96 angstroms. These structures reveal a protective mechanism by which two domains of NS1 are antagonized simultaneously. The NS1 wing domain mediates cell binding, whereas the β-ladder triggers downstream events, both of which are required for dengue, Zika, and West Nile virus NS1-mediated endothelial dysfunction. These observations provide a mechanistic explanation for 2B7 protection against NS1-induced pathology and demonstrate the potential of one antibody to treat infections by multiple flaviviruses.
Copyright © 2021, American Association for the Advancement of Science.
Conflict of interest statement
Figures


Comment in
-
How NS1 Antibodies Prevent Severe Flavivirus Disease.Trends Biochem Sci. 2021 Jul;46(7):519-521. doi: 10.1016/j.tibs.2021.03.005. Epub 2021 Apr 22. Trends Biochem Sci. 2021. PMID: 33895084
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

