U2af1 is required for survival and function of hematopoietic stem/progenitor cells
- PMID: 33414485
- PMCID: PMC8283943
- DOI: 10.1038/s41375-020-01116-x
U2af1 is required for survival and function of hematopoietic stem/progenitor cells
Abstract
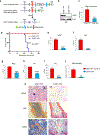
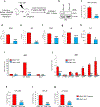
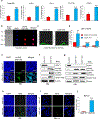
U2AF1 is involved in the recognition of the 3' splice site during pre-mRNA splicing. Mutations in U2AF1 are frequently observed in myelodysplastic syndromes. However, the role of wild-type U2AF1 in normal hematopoiesis has remained elusive. Using a novel conditional U2af1 knockout allele, we have found that deletion of U2af1 results in profound defects in hematopoiesis characterized by pancytopenia, ablation of hematopoietic stem/progenitor cells (HSPC) leading to bone marrow failure and early lethality in mice. U2af1 deletion impairs HSPC function and repopulation capacity. U2af1 deletion also causes increased DNA damage and reduced survival in hematopoietic progenitors. RNA sequencing analysis reveals significant alterations in the expression of genes related to HSC maintenance, cell proliferation, and DNA damage response-related pathways in U2af1-deficient HSPC. U2af1 deficiency also induces splicing alterations in genes important for HSPC function. This includes altered splicing and perturbed expression of Nfya and Pbx1 transcription factors in U2af1-deficient HSPC. Collectively, these results suggest an important role for U2af1 in the maintenance and function of HSPC in normal hematopoiesis. A better understanding of the normal function of U2AF1 in hematopoiesis is important for development of appropriate therapeutic approaches for U2AF1 mutant induced hematologic malignancies.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited part of Springer Nature.
Conflict of interest statement
Conflict of interest
The authors declare that they have no conflict of interest.
Figures




References
-
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent Pathway Mutations of Splicing Machinery in Myelodysplasia. Nature, 2011. 478(7367):64–69. - PubMed
-
- Thol F, Kade S, Schlarmann C, Löffeld P, Morgan M, Krauter J, et al. Frequency and Prognostic Impact of Mutations in SRSF2, U2AF1, and ZRSR2 in Patients With Myelodysplastic Syndromes. Blood, 2012. 119(15): 3578–84. - PubMed
-
- Damm F, Kosmider O, Gelsi-Boyer V, Renneville A, Carbuccia N, Hidalgo-Curtis C, et al. Groupe Francophone des Myélodysplasies., Mutations Affecting mRNA Splicing Define Distinct Clinical Phenotypes and Correlate With Patient Outcome in Myelodysplastic Syndromes. Blood, 2012. 119(14): 3211–18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

