Investigating the genetic architecture of noncognitive skills using GWAS-by-subtraction
- PMID: 33414549
- PMCID: PMC7116735
- DOI: 10.1038/s41588-020-00754-2
Investigating the genetic architecture of noncognitive skills using GWAS-by-subtraction
Abstract
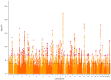
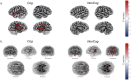
Little is known about the genetic architecture of traits affecting educational attainment other than cognitive ability. We used genomic structural equation modeling and prior genome-wide association studies (GWASs) of educational attainment (n = 1,131,881) and cognitive test performance (n = 257,841) to estimate SNP associations with educational attainment variation that is independent of cognitive ability. We identified 157 genome-wide-significant loci and a polygenic architecture accounting for 57% of genetic variance in educational attainment. Noncognitive genetics were enriched in the same brain tissues and cell types as cognitive performance, but showed different associations with gray-matter brain volumes. Noncognitive genetics were further distinguished by associations with personality traits, less risky behavior and increased risk for certain psychiatric disorders. For socioeconomic success and longevity, noncognitive and cognitive-performance genetics demonstrated associations of similar magnitude. By conducting a GWAS of a phenotype that was not directly measured, we offer a view of genetic architecture of noncognitive skills influencing educational success.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- von Stumm S, Hell B, Chamorro-Premuzic T. The hungry mind: intellectual curiosity is the third pillar of academic performance. Perspect Psychol Sci. 2011;6:574–588. - PubMed
-
- Heckman JJ, Stixrud J, Urzua S. The effects of cognitive and noncognitive abilities on labor market outcomes and social behavior. J Labor Econ. 2006;24:411–482.
Publication types
MeSH terms
Grants and funding
- RC2 MH089951/MH/NIMH NIH HHS/United States
- MR/P005918/1/MRC_/Medical Research Council/United Kingdom
- R01 HD077482/HD/NICHD NIH HHS/United States
- R01 HD073342/HD/NICHD NIH HHS/United States
- R01 AG054628/AG/NIA NIH HHS/United States
- P2C HD042849/HD/NICHD NIH HHS/United States
- 230374/ERC_/European Research Council/International
- EP-C-15-001/EPA/EPA/United States
- MR/R024065/1/MRC_/Medical Research Council/United Kingdom
- G1002190/MRC_/Medical Research Council/United Kingdom
- R01 HD092548/HD/NICHD NIH HHS/United States
- R01 AG041868/AG/NIA NIH HHS/United States
- P01 HD031921/HD/NICHD NIH HHS/United States
- RC2 MH089995/MH/NIMH NIH HHS/United States
- R01 HD060726/HD/NICHD NIH HHS/United States
- P30 AG017266/AG/NIA NIH HHS/United States
- R01 MD011716/MD/NIMHD NIH HHS/United States
- R01 HD083613/HD/NICHD NIH HHS/United States
- R24 HD042849/HD/NICHD NIH HHS/United States
- R01 AG032282/AG/NIA NIH HHS/United States
- R01 MH120219/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

