Conservation of copy number profiles during engraftment and passaging of patient-derived cancer xenografts
- PMID: 33414553
- PMCID: PMC7808565
- DOI: 10.1038/s41588-020-00750-6
Conservation of copy number profiles during engraftment and passaging of patient-derived cancer xenografts
Erratum in
-
Author Correction: Conservation of copy number profiles during engraftment and passaging of patient-derived cancer xenografts.Nat Genet. 2021 May;53(5):761. doi: 10.1038/s41588-021-00811-4. Nat Genet. 2021. PMID: 33608693 Free PMC article. No abstract available.
Abstract
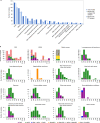
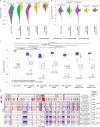
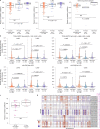
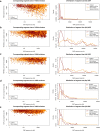
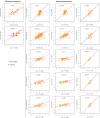
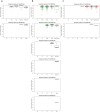
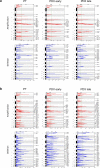
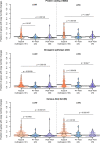
Patient-derived xenografts (PDXs) are resected human tumors engrafted into mice for preclinical studies and therapeutic testing. It has been proposed that the mouse host affects tumor evolution during PDX engraftment and propagation, affecting the accuracy of PDX modeling of human cancer. Here, we exhaustively analyze copy number alterations (CNAs) in 1,451 PDX and matched patient tumor (PT) samples from 509 PDX models. CNA inferences based on DNA sequencing and microarray data displayed substantially higher resolution and dynamic range than gene expression-based inferences, and they also showed strong CNA conservation from PTs through late-passage PDXs. CNA recurrence analysis of 130 colorectal and breast PT/PDX-early/PDX-late trios confirmed high-resolution CNA retention. We observed no significant enrichment of cancer-related genes in PDX-specific CNAs across models. Moreover, CNA differences between patient and PDX tumors were comparable to variations in multiregion samples within patients. Our study demonstrates the lack of systematic copy number evolution driven by the PDX mouse host.
Conflict of interest statement
A.L.W. and B.E.W. receive a portion of royalties if the University of Utah licenses certain PDX models to for-profit entities. M.T.L. is a founder of, and equity stake holder in, Tvardi Therapeutics, a founder of, and limited partner in, StemMed and a manager in StemMed Holdings. He also receives a portion of royalties if the Baylor College of Medicine licenses certain PDX models to for-profit entities. J.A.R. serves as a consultant and received stocks from Genprex, and receives royalties from patents issued. F.M.-B. reports receiving commercial research grants from Novartis, AstraZeneca, Calithera, Aileron, Bayer, Jounce, CytomX, eFFECTOR, Zymeworks, PUMA Biotechnology, Curis, Millennium, Daiichi Sankyo, Abbvie, Guardant Health, Takeda, Seattle Genetics and GlaxoSmithKline, as well as grants and travel-related fees from Taiho, Genentech, Debiopharm Group and Pfizer. She also served as a consultant to Pieris, Dialectica, Sumitomo Dainippon, Samsung Bioepis, Aduro, OrigiMed, Xencor, The Jackson Laboratory, Zymeworks, Kolon Life Science and Parexel International, and an advisor to Inflection Biosciences, GRAIL, DarwinHealth, Spectrum, Mersana and Seattle Genetics. L.T. reports receiving research grants from Symphogen, Servier, Pfizer and Merus, and he is in the speakers’ bureau of Eli Lilly, AstraZeneca and Merck. J.J. reports receiving funding for collaborative research from Artios Pharma. He also serves as a Scientific Advisory Board member of Artios Pharma. The other authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- U54 CA233223/CA/NCI NIH HHS/United States
- P30 CA034196/CA/NCI NIH HHS/United States
- P01 CA114046/CA/NCI NIH HHS/United States
- T32 HG008962/HG/NHGRI NIH HHS/United States
- HHSN261201400008C/CA/NCI NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- U24 CA224067/CA/NCI NIH HHS/United States
- P50 CA196510/CA/NCI NIH HHS/United States
- U54 CA224070/CA/NCI NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- 29567/CRUK_/Cancer Research UK/United Kingdom
- U54 CA224076/CA/NCI NIH HHS/United States
- U54 CA224065/CA/NCI NIH HHS/United States
- U54 CA233306/CA/NCI NIH HHS/United States
- P30 CA010815/CA/NCI NIH HHS/United States
- U24 CA204781/CA/NCI NIH HHS/United States
- U54 CA224083/CA/NCI NIH HHS/United States
- HHSN261201500003C/CA/NCI NIH HHS/United States
- R50 CA211199/CA/NCI NIH HHS/United States
- P30 CA125123/CA/NCI NIH HHS/United States
- NC/T001267/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- P50 CA070907/CA/NCI NIH HHS/United States
- HHSN261201500003I/CA/NCI NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- P30 CA042014/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

