Targeting PAI-1 in Cardiovascular Disease: Structural Insights Into PAI-1 Functionality and Inhibition
- PMID: 33415130
- PMCID: PMC7782431
- DOI: 10.3389/fcvm.2020.622473
Targeting PAI-1 in Cardiovascular Disease: Structural Insights Into PAI-1 Functionality and Inhibition
Abstract
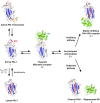
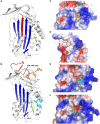
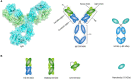
Plasminogen activator inhibitor-1 (PAI-1), a member of the serine protease inhibitor (serpin) superfamily with antiprotease activity, is the main physiological inhibitor of tissue-type (tPA) and urokinase-type (uPA) plasminogen activators (PAs). Apart from being crucially involved in fibrinolysis and wound healing, PAI-1 plays a pivotal role in various acute and chronic pathophysiological processes, including cardiovascular disease, tissue fibrosis, cancer, and age-related diseases. In the prospect of treating the broad range of PAI-1-related pathologies, many efforts have been devoted to developing PAI-1 inhibitors. The use of these inhibitors, including low molecular weight molecules, peptides, antibodies, and antibody fragments, in various animal disease models has provided ample evidence of their beneficial effect in vivo and moved forward some of these inhibitors in clinical trials. However, none of these inhibitors is currently approved for therapeutic use in humans, mainly due to selectivity and toxicity issues. Furthermore, the conformational plasticity of PAI-1, which is unique among serpins, poses a real challenge in the identification and development of PAI-1 inhibitors. This review will provide an overview of the structural insights into PAI-1 functionality and modulation thereof and will highlight diverse approaches to inhibit PAI-1 activity.
Keywords: PAI-1 inhibitors; cardiovascular disease; fibrinolyisis; plasminogen activator inhibitor 1 (PAI-1); serpin (serine proteinase inhibitor).
Copyright © 2020 Sillen and Declerck.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Small molecules inhibitors of plasminogen activator inhibitor-1 - an overview.Eur J Med Chem. 2015 Mar 6;92:619-36. doi: 10.1016/j.ejmech.2015.01.010. Epub 2015 Jan 8. Eur J Med Chem. 2015. PMID: 25615797 Review.
-
Structural Insights into the Mechanism of a Nanobody That Stabilizes PAI-1 and Modulates Its Activity.Int J Mol Sci. 2020 Aug 15;21(16):5859. doi: 10.3390/ijms21165859. Int J Mol Sci. 2020. PMID: 32824134 Free PMC article.
-
Molecular mechanism of two nanobodies that inhibit PAI-1 activity reveals a modulation at distinct stages of the PAI-1/plasminogen activator interaction.J Thromb Haemost. 2020 Mar;18(3):681-692. doi: 10.1111/jth.14716. Epub 2020 Feb 20. J Thromb Haemost. 2020. PMID: 31858714 Free PMC article.
-
Beyond fibrinolysis: the role of plasminogen activator inhibitor-1 and vitronectin in vascular wound healing.Trends Cardiovasc Med. 1998 May;8(4):175-80. doi: 10.1016/S1050-1738(98)00003-6. Trends Cardiovasc Med. 1998. PMID: 21235930
-
Serpin regulation of fibrinolytic system: implications for therapeutic applications in cardiovascular diseases.Cardiovasc Hematol Agents Med Chem. 2014;12(2):91-125. doi: 10.2174/1871525712666141106095927. Cardiovasc Hematol Agents Med Chem. 2014. PMID: 25374013 Review.
Cited by
-
A Narrative Review on Plasminogen Activator Inhibitor-1 and Its (Patho)Physiological Role: To Target or Not to Target?Int J Mol Sci. 2021 Mar 8;22(5):2721. doi: 10.3390/ijms22052721. Int J Mol Sci. 2021. PMID: 33800359 Free PMC article. Review.
-
Ligand-mediated PAI-1 inhibition in a mouse model of peritoneal carcinomatosis.Cell Rep Med. 2022 Feb 15;3(2):100526. doi: 10.1016/j.xcrm.2022.100526. eCollection 2022 Feb 15. Cell Rep Med. 2022. PMID: 35243423 Free PMC article.
-
Plasminogen activator inhibitor-1 promoter sequence variations in idiopathic osteonecrosis of head of femur.Indian J Med Res. 2021 Jun;154(6):849-856. doi: 10.4103/ijmr.IJMR_496_19. Indian J Med Res. 2021. PMID: 35662090 Free PMC article.
-
PAI-1 Regulates the Cytoskeleton and Intrinsic Stiffness of Vascular Smooth Muscle Cells.Arterioscler Thromb Vasc Biol. 2024 Oct;44(10):2191-2203. doi: 10.1161/ATVBAHA.124.320938. Epub 2024 Jun 13. Arterioscler Thromb Vasc Biol. 2024. PMID: 38868940
-
Irisin as a Novel Biomarker of Subclinical Atherosclerosis in Severe Obesity.Int J Mol Sci. 2023 May 3;24(9):8171. doi: 10.3390/ijms24098171. Int J Mol Sci. 2023. PMID: 37175880 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

