Identification of Novel Long Noncoding RNAs and Their Role in Abdominal Aortic Aneurysm
- PMID: 33415145
- PMCID: PMC7769652
- DOI: 10.1155/2020/3502518
Identification of Novel Long Noncoding RNAs and Their Role in Abdominal Aortic Aneurysm
Abstract
Objective: Long noncoding RNAs (lncRNAs) have emerged as critical molecular regulators in various diseases. However, the potential regulatory role of lncRNAs in the pathogenesis of abdominal aortic aneurysm (AAA) remains elusive. The aim of this study was to identify crucial lncRNAs associated with human AAA by comparing the lncRNA and mRNA expression profiles of patients with AAA with those of control individuals.
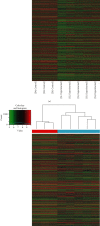
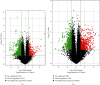
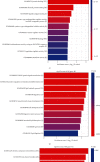
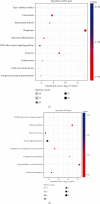
Materials and methods: The expression profiles of lncRNAs and mRNAs were analyzed in five dilated aortic samples from AAA patients and three normal aortic samples from control individuals using microarray technology. Functional annotation of the screened lncRNAs based on the differentially expressed genes was performed using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses.
Results: Microarray results revealed 2046 lncRNAs and 1363 mRNAs. Functional enrichment analysis showed that the mRNAs significantly associated with AAA were enriched in the NOD-like receptor (NLR) and nuclear factor kappa-B (NF-κB) signaling pathways and in cell adhesion molecules (CAMs), which are closely associated with pathophysiological changes in AAA. The lncRNAs identified using microarray analysis were further validated using quantitative real-time polymerase chain reaction (qRT-PCR) analysis with 12 versus 11 aortic samples. Finally, three key lncRNAs (ENST00000566954, ENST00000580897, and T181556) were confirmed using strict validation. A coding-noncoding coexpression (CNC) network and a competing endogenous RNA (ceRNA) network were constructed to determine the interaction among the lncRNAs, microRNAs, and mRNAs based on the confirmed lncRNAs.
Conclusions: Our microarray profiling analysis and validation of significantly expressed lncRNAs between patients with AAA and control group individuals may provide new diagnostic biomarkers for AAA. The underlying regulatory mechanisms of the confirmed lncRNAs in AAA pathogenesis need to be determined using in vitro and in vivo experiments.
Copyright © 2020 Abulaihaiti Maitiseyiti et al.
Conflict of interest statement
The authors declare no conflicts of interest, financial, or otherwise.
Figures



Similar articles
-
Long Noncoding RNA Expression Signatures of Abdominal Aortic Aneurysm Revealed by Microarray.Biomed Environ Sci. 2016 Oct;29(10):713-723. doi: 10.3967/bes2016.096. Biomed Environ Sci. 2016. PMID: 27927271
-
Differentially-expressed mRNAs, microRNAs and long noncoding RNAs in intervertebral disc degeneration identified by RNA-sequencing.Bioengineered. 2021 Dec;12(1):1026-1039. doi: 10.1080/21655979.2021.1899533. Bioengineered. 2021. PMID: 33764282 Free PMC article.
-
Identification of lncRNA and mRNA Biomarkers in Osteoarthritic Degenerative Meniscus by Weighted Gene Coexpression Network and Competing Endogenous RNA Network Analysis.Biomed Res Int. 2020 May 20;2020:2123787. doi: 10.1155/2020/2123787. eCollection 2020. Biomed Res Int. 2020. PMID: 32685450 Free PMC article.
-
Pathogenetic Significance of Long Non-Coding RNAs in the Development of Thoracic and Abdominal Aortic Aneurysms.Biochemistry (Mosc). 2024 Jan;89(1):130-147. doi: 10.1134/S0006297924010085. Biochemistry (Mosc). 2024. PMID: 38467550 Review.
-
Identification and bioinformatics analysis of lncRNAs in serum of patients with ankylosing spondylitis.BMC Musculoskelet Disord. 2024 Apr 15;25(1):291. doi: 10.1186/s12891-024-07396-z. BMC Musculoskelet Disord. 2024. PMID: 38622662 Free PMC article. Review.
Cited by
-
Structural and Pharmacological Network Analysis of miRNAs Involved in Acute Ischemic Stroke: A Systematic Review.Int J Mol Sci. 2022 Apr 23;23(9):4663. doi: 10.3390/ijms23094663. Int J Mol Sci. 2022. PMID: 35563054 Free PMC article.
-
Identification of endoplasmic reticulum stress-associated lncRNAs influencing inflammation and VSMC function in abdominal aortic aneurysm.Clin Sci (Lond). 2025 Mar 25;139(6):357-72. doi: 10.1042/CS20242476. Clin Sci (Lond). 2025. PMID: 40072504 Free PMC article.
-
Biomarkers in EndoVascular Aneurysm Repair (EVAR) and Abdominal Aortic Aneurysm: Pathophysiology and Clinical Implications.Diagnostics (Basel). 2022 Jan 13;12(1):183. doi: 10.3390/diagnostics12010183. Diagnostics (Basel). 2022. PMID: 35054350 Free PMC article. Review.
-
Key ferroptosis-related genes in abdominal aortic aneurysm formation and rupture as determined by combining bioinformatics techniques.Front Cardiovasc Med. 2022 Aug 9;9:875434. doi: 10.3389/fcvm.2022.875434. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 36017103 Free PMC article.
References
-
- Sakalihasan N., Limet R., Defawe O. D. Abdominal aortic aneurysm. Lancet. 2005;365(9470):1577–1589. - PubMed
-
- Kent K. C. Clinical practice. Abdominal aortic aneurysms. New England Journal of Medicine. 2014;371(22):2101–2108. - PubMed
-
- Davis F. M., Rateri D. L., Daugherty A. Mechanisms of aortic aneurysm formation: translating preclinical studies into clinical therapies. Heart. 2014;100(19):1498–1505. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

