Unsupervised cluster analysis of SARS-CoV-2 genomes reflects its geographic progression and identifies distinct genetic subgroups of SARS-CoV-2 virus
- PMID: 33415739
- PMCID: PMC8005425
- DOI: 10.1002/gepi.22373
Unsupervised cluster analysis of SARS-CoV-2 genomes reflects its geographic progression and identifies distinct genetic subgroups of SARS-CoV-2 virus
Abstract
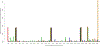
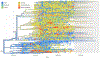
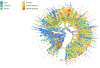
Over 10,000 viral genome sequences of the SARS-CoV-2virus have been made readily available during the ongoing coronavirus pandemic since the initial genome sequence of the virus was released on the open access Virological website (http://virological.org/) early on January 11. We utilize the published data on the single stranded RNAs of 11,132 SARS-CoV-2 patients in the GISAID database, which contains fully or partially sequenced SARS-CoV-2 samples from laboratories around the world. Among many important research questions which are currently being investigated, one aspect pertains to the genetic characterization/classification of the virus. We analyze data on the nucleotide sequencing of the virus and geographic information of a subset of 7640 SARS-CoV-2 patients without missing entries that are available in the GISAID database. Instead of modeling the mutation rate, applying phylogenetic tree approaches, and so forth, we here utilize a model-free clustering approach that compares the viruses at a genome-wide level. We apply principal component analysis to a similarity matrix that compares all pairs of these SARS-CoV-2 nucleotide sequences at all loci simultaneously, using the Jaccard index. Our analysis results of the SARS-CoV-2 genome data illustrates the geographic and chronological progression of the virus, starting from the first cases that were observed in China to the current wave of cases in Europe and North America. This is in line with a phylogenetic analysis which we use to contrast our results. We also observe that, based on their sequence data, the SARS-CoV-2 viruses cluster in distinct genetic subgroups. It is the subject of ongoing research to examine whether the genetic subgroup could be related to diseases outcome and its potential implications for vaccine development.
Keywords: SARS-CoV-2; clustering; covid; jaccard.
© 2021 Wiley Periodicals LLC.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures
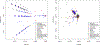
Update of
-
Unsupervised cluster analysis of SARS-CoV-2 genomes reflects its geographic progression and identifies distinct genetic subgroups of SARS-CoV-2 virus.bioRxiv [Preprint]. 2020 Nov 20:2020.05.05.079061. doi: 10.1101/2020.05.05.079061. bioRxiv. 2020. Update in: Genet Epidemiol. 2021 Apr;45(3):316-323. doi: 10.1002/gepi.22373. PMID: 32637949 Free PMC article. Updated. Preprint.
References
-
- Freunde of GISAID, e. (2020). Global Initiative on Sharing All Influenza Data – TreeTool App.
-
- Hahn G, Cho MH, Weiss ST, Silverman EK, and Lange C (2020a). Unsupervised cluster analysis of SARS-CoV-2 genomes indicates that recent (June 2020) cases in Beijing are from a genetic subgroup that consists of mostly European and South (east) Asian samples, of which the latter are the most recent. bioRxiv:2020.06.22.165936.
-
- Hahn G, Lutz S, and Lange C (2020c). locStra: Fast Implementation of (Local) Population Stratification Methods (v1.3) https://cran.r-project.org/web/packages/locStra/index.html.
Publication types
MeSH terms
Grants and funding
- U01HL089897/HL/NHLBI NIH HHS/United States
- P01HL132825/HL/NHLBI NIH HHS/United States
- R01 AI154470/AI/NIAID NIH HHS/United States
- R01HG008976/HG/NHGRI NIH HHS/United States
- P01HL120839/HL/NHLBI NIH HHS/United States
- R01 HG008976/HG/NHGRI NIH HHS/United States
- Cure Alzheimer's Fund
- 2U01HG008685/HL/NHLBI NIH HHS/United States
- U01 HG008685/HG/NHGRI NIH HHS/United States
- P01 HL132825/HL/NHLBI NIH HHS/United States
- U01HL089856/HL/NHLBI NIH HHS/United States
- U01 HL089897/HL/NHLBI NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- National Institutes of Health: 1R01AI154470-01; 2U01HG008685
- P01 HL120839/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

