Heterogeneous disease-propagating stem cells in juvenile myelomonocytic leukemia
- PMID: 33416891
- PMCID: PMC7802370
- DOI: 10.1084/jem.20180853
Heterogeneous disease-propagating stem cells in juvenile myelomonocytic leukemia
Abstract
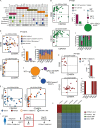
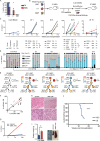
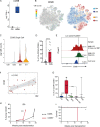
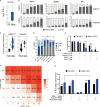
Juvenile myelomonocytic leukemia (JMML) is a poor-prognosis childhood leukemia usually caused by RAS-pathway mutations. The cellular hierarchy in JMML is poorly characterized, including the identity of leukemia stem cells (LSCs). FACS and single-cell RNA sequencing reveal marked heterogeneity of JMML hematopoietic stem/progenitor cells (HSPCs), including an aberrant Lin-CD34+CD38-CD90+CD45RA+ population. Single-cell HSPC index-sorting and clonogenic assays show that (1) all somatic mutations can be backtracked to the phenotypic HSC compartment, with RAS-pathway mutations as a "first hit," (2) mutations are acquired with both linear and branching patterns of clonal evolution, and (3) mutant HSPCs are present after allogeneic HSC transplant before molecular/clinical evidence of relapse. Stem cell assays reveal interpatient heterogeneity of JMML LSCs, which are present in, but not confined to, the phenotypic HSC compartment. RNA sequencing of JMML LSC reveals up-regulation of stem cell and fetal genes (HLF, MEIS1, CNN3, VNN2, and HMGA2) and candidate therapeutic targets/biomarkers (MTOR, SLC2A1, and CD96), paving the way for LSC-directed disease monitoring and therapy in this disease.
© 2021 Louka et al.
Conflict of interest statement
Disclosures: S. O’Byrne reports personal fees from Becton, Dickinson and Company (employee as of August 2020) outside the submitted work. M. Salio reports personal fees from Nucleome Therapeutics outside the submitted work. N. Sousos reports expenses reimbursement from Pfizer Limited for attending the Pfizer-organized meeting titled "Leaders in Leukaemia," July 12-13, 2019, in London, UK. N. Sousos reports expenses reimbursement from Constellation Pharmaceuticals for attending the Constellation 0610-02 MANIFEST Study EU Investigator Meeting, January 31-February 1, 2019, in Rome, Italy. N. Sousos reports an educational travel grant from AOP Orphan Pharmaceuticals AG for attending the European Hematology Association meeting "Diagnosis and Management of Myeloproliferative Neoplasms," October 12-14, 2017, in Budapest, Hungary. No other disclosures were reported.
Figures








Comment in
-
Stem cell origins of JMML.J Exp Med. 2021 Feb 1;218(2):e20202152. doi: 10.1084/jem.20202152. J Exp Med. 2021. PMID: 33427877 Free PMC article.
References
-
- Caye, A., Rouault-Pierre K., Strullu M., Lainey E., Abarrategi A., Fenneteau O., Arfeuille C., Osman J., Cassinat B., Pereira S., et al. . 2020. Despite mutation acquisition in hematopoietic stem cells, JMML-propagating cells are not always restricted to this compartment. Leukemia. 34:1658–1668. 10.1038/s41375-019-0662-y - DOI - PMC - PubMed
-
- Chávez-González, A., Dorantes-Acosta E., Moreno-Lorenzana D., Alvarado-Moreno A., Arriaga-Pizano L., and Mayani H.. 2014. Expression of CD90, CD96, CD117, and CD123 on different hematopoietic cell populations from pediatric patients with acute myeloid leukemia. Arch. Med. Res. 45:343–350. 10.1016/j.arcmed.2014.04.001 - DOI - PubMed
-
- Dimitriou, M., Woll P.S., Mortera-Blanco T., Karimi M., Wedge D.C., Doolittle H., Douagi I., Papaemmanuil E., Jacobsen S.E.W., and Hellström-Lindberg E.. 2016. Perturbed hematopoietic stem and progenitor cell hierarchy in myelodysplastic syndromes patients with monosomy 7 as the sole cytogenetic abnormality. Oncotarget. 7:72685–72698. 10.18632/oncotarget.12234 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

