Molecular Pathogenesis and Treatment Perspectives for Hypereosinophilia and Hypereosinophilic Syndromes
- PMID: 33418988
- PMCID: PMC7825323
- DOI: 10.3390/ijms22020486
Molecular Pathogenesis and Treatment Perspectives for Hypereosinophilia and Hypereosinophilic Syndromes
Abstract
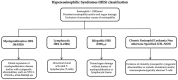
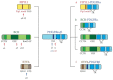
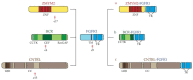
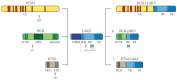
Hypereosinophilia (HE) is a heterogeneous condition with a persistent elevated eosinophil count of >350/mm3, which is reported in various (inflammatory, allergic, infectious, or neoplastic) diseases with distinct pathophysiological pathways. HE may be associated with tissue or organ damage and, in this case, the disorder is classified as hypereosinophilic syndrome (HES). Different studies have allowed for the discovery of two major pathogenetic variants known as myeloid or lymphocytic HES. With the advent of molecular genetic analyses, such as T-cell receptor gene rearrangement assays and Next Generation Sequencing, it is possible to better characterize these syndromes and establish which patients will benefit from pharmacological targeted therapy. In this review, we highlight the molecular alterations that are involved in the pathogenesis of eosinophil disorders and revise possible therapeutic approaches, either implemented in clinical practice or currently under investigation in clinical trials.
Keywords: NGS; PDGFRα and PDGFRβ fusions; TCR rearrangements; hypereosinophilia; hypereosinophilic syndromes.
Conflict of interest statement
Giuseppe Alberto Palumbo: homoraria from Novartis, Bristol Myers Squibb, Celgen, Amgen and Abbvie Paolo Vigneri: research funding from Novartis and Pfizer; honoraria from Astra-Zeneca, Celgene, Italfarmaco, Incyte, Novartis, Pfizer, Tesaro, and Teva. The other authors declare no conflict of interest.
Figures
Similar articles
-
Management of Hypereosinophilic Syndromes.Immunol Allergy Clin North Am. 2015 Aug;35(3):561-75. doi: 10.1016/j.iac.2015.05.006. Immunol Allergy Clin North Am. 2015. PMID: 26209900 Review.
-
Molecular classification and pathogenesis of eosinophilic disorders: 2005 update.Acta Haematol. 2005;114(1):7-25. doi: 10.1159/000085559. Acta Haematol. 2005. PMID: 15995322 Review.
-
Biologic therapies for hypereosinophilic disorders: From tyrosine kinase inhibitors to monoclonal antibodies. Towards an increasingly customized management?Blood Rev. 2023 Mar;58:101014. doi: 10.1016/j.blre.2022.101014. Epub 2022 Sep 17. Blood Rev. 2023. PMID: 36153195 Review.
-
[New insights into hypereosinophilic syndromes].Bull Acad Natl Med. 2010 Mar;194(3):547-59; discussion 559-60. Bull Acad Natl Med. 2010. PMID: 21171248 French.
-
Imatinib for the treatment of hypereosinophilic syndromes.Expert Rev Clin Immunol. 2018 Feb;14(2):163-170. doi: 10.1080/1744666X.2018.1425142. Epub 2018 Jan 9. Expert Rev Clin Immunol. 2018. PMID: 29303368 Review.
Cited by
-
Perioperative Respiratory Outcome of Patients with Eosinophilia: A Cohort Study in a Tertiary Care Hospital.Anesthesiol Res Pract. 2023 Jan 10;2023:8514949. doi: 10.1155/2023/8514949. eCollection 2023. Anesthesiol Res Pract. 2023. PMID: 36660020 Free PMC article.
-
Clinical analysis of hypereosinophilic syndrome first presenting with asthma-like symptoms.Ann Med. 2022 Dec;54(1):11-21. doi: 10.1080/07853890.2021.2014555. Ann Med. 2022. PMID: 34935570 Free PMC article.
-
Management of eosinophil-associated inflammatory diseases: the importance of a multidisciplinary approach.Front Immunol. 2023 May 17;14:1192284. doi: 10.3389/fimmu.2023.1192284. eCollection 2023. Front Immunol. 2023. PMID: 37266434 Free PMC article.
-
The multidisciplinary approach to eosinophilia.Front Oncol. 2023 May 18;13:1193730. doi: 10.3389/fonc.2023.1193730. eCollection 2023. Front Oncol. 2023. PMID: 37274287 Free PMC article. Review.
-
Mepolizumab in Hypereosinophilic Syndrome: A Systematic Review and Meta-analysis.Clinics (Sao Paulo). 2021 Oct 11;76:e3271. doi: 10.6061/clinics/2021/e3271. eCollection 2021. Clinics (Sao Paulo). 2021. PMID: 34644737 Free PMC article.
References
-
- Dvorak A.M., Letourneau L., Login G.R., Weller P.F., Ackerman S.J. Ultrastructural localization of the Charcot-Leyden crystal protein (lysophospholipase) to a distinct crystalloid-free granule population in mature human eosinophils. Blood. 1988;72:150–158. doi: 10.1182/blood.V72.1.150.150. - DOI - PubMed
-
- Giembycz M.A., Lindsay M.A. Pharmacology of the eosinophil. Pharmacol. Rev. 1999;51:213–340. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

