Structural Characterization of Act c 10.0101 and Pun g 1.0101-Allergens from the Non-Specific Lipid Transfer Protein Family
- PMID: 33419110
- PMCID: PMC7825401
- DOI: 10.3390/molecules26020256
Structural Characterization of Act c 10.0101 and Pun g 1.0101-Allergens from the Non-Specific Lipid Transfer Protein Family
Abstract
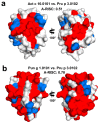
(1) Background: Non-specific lipid transfer proteins (nsLTPs), which belong to the prolamin superfamily, are potent allergens. While the biological role of LTPs is still not well understood, it is known that these proteins bind lipids. Allergen nsLTPs are characterized by significant stability and resistance to digestion. (2) Methods: nsLTPs from gold kiwifruit (Act c 10.0101) and pomegranate (Pun g 1.0101) were isolated from their natural sources and structurally characterized using X-ray crystallography (3) Results: Both proteins crystallized and their crystal structures were determined. The proteins have a very similar overall fold with characteristic compact, mainly α-helical structures. The C-terminal sequence of Act c 10.0101 was updated based on our structural and mass spectrometry analysis. Information on proteins' sequences and structures was used to estimate the risk of cross-reactive reactions between Act c 10.0101 or Pun g 1.0101 and other allergens from this family of proteins. (4) Conclusions: Structural studies indicate a conformational flexibility of allergens from the nsLTP family and suggest that immunoglobulin E binding to some surface regions of these allergens may depend on ligand binding. Both Act c 10.0101 and Pun g 1.0101 are likely to be involved in cross-reactive reactions involving other proteins from the nsLTP family.
Keywords: cross-reactivity; food allergy; natural source; nsLTP; structure.
Conflict of interest statement
A.O., S.P., A.G.-C., and M.C. declare no conflicts of interest. K.K. received lecture fees from: ALK Abello, Astra Zeneca, Berlin Chemie, Chiesi, Emma, Hal Allergy, Meda Pharma, Orion Pharma, and royalties from UpToDate. I.G. and L.T. are employees of ADL srl, and M.A.C. receives funding from ADL srl.
Figures

Similar articles
-
Pomegranate ( Punica granatum L.) expresses several nsLTP isoforms characterized by different immunoglobulin E-binding properties.Int Arch Allergy Immunol. 2014;164(2):112-21. doi: 10.1159/000362761. Epub 2014 Jun 17. Int Arch Allergy Immunol. 2014. PMID: 24941918
-
Structural and Functional Characterization of the Hazelnut Allergen Cor a 8.J Agric Food Chem. 2015 Oct 21;63(41):9150-8. doi: 10.1021/acs.jafc.5b03534. Epub 2015 Oct 7. J Agric Food Chem. 2015. PMID: 26417906 Free PMC article.
-
Cysteine proteinase inhibitor Act d 4 is a functional allergen contributing to the clinical symptoms of kiwifruit allergy.Mol Nutr Food Res. 2010 Mar;54(3):373-80. doi: 10.1002/mnfr.200900035. Mol Nutr Food Res. 2010. PMID: 19885843
-
Interaction of Non-Specific Lipid-Transfer Proteins With Plant-Derived Lipids and Its Impact on Allergic Sensitization.Front Immunol. 2018 Jun 20;9:1389. doi: 10.3389/fimmu.2018.01389. eCollection 2018. Front Immunol. 2018. PMID: 29973934 Free PMC article. Review.
-
Plant non-specific lipid transfer proteins: an interface between plant defence and human allergy.Biochim Biophys Acta. 2007 Jun;1771(6):781-91. doi: 10.1016/j.bbalip.2007.01.001. Epub 2007 Jan 8. Biochim Biophys Acta. 2007. PMID: 17349819 Review.
Cited by
-
Ultrasound and Heat Treatment and Its Potential to Reduce Fennel Allergenicity.Foods. 2025 Jun 25;14(13):2251. doi: 10.3390/foods14132251. Foods. 2025. PMID: 40647003 Free PMC article.
-
Structural Basis of the Immunological Cross-Reactivity between Kiwi and Birch Pollen.Foods. 2023 Oct 27;12(21):3939. doi: 10.3390/foods12213939. Foods. 2023. PMID: 37959058 Free PMC article.
-
Role of Small Molecule Ligands in IgE-Mediated Allergy.Curr Allergy Asthma Rep. 2023 Sep;23(9):497-508. doi: 10.1007/s11882-023-01100-2. Epub 2023 Jun 23. Curr Allergy Asthma Rep. 2023. PMID: 37351723 Free PMC article. Review.
-
Isolation, Characterization and IgE Binding of Two 2S Albumins of Pomegranate Seeds.Foods. 2024 Jun 21;13(13):1965. doi: 10.3390/foods13131965. Foods. 2024. PMID: 38998471 Free PMC article.
-
When the Frequencies of Sensitization and Elicitation of Allergic Reaction Do Not Correlate-The Case of Apple Gibberellin-Regulated Protein Tested in an Italian Population.Front Allergy. 2021 Oct 21;2:745825. doi: 10.3389/falgy.2021.745825. eCollection 2021. Front Allergy. 2021. PMID: 35387024 Free PMC article.
References
-
- Pasquato N., Berni R., Folli C., Folloni S., Cianci M., Pantano S., Helliwell J.R., Zanotti G. Crystal structure of peach Pru p 3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens. J. Mol. Biol. 2006;356:684–694. doi: 10.1016/j.jmb.2005.11.063. - DOI - PubMed
-
- Bogdanov I.V., Shenkarev Z.O., Finkina E.I., Melnikova D.N., Rumynskiy E.I., Arseniev A.S., Ovchinnikova T.V. A novel lipid transfer protein from the pea Pisum sativum: Isolation, recombinant expression, solution structure, antifungal activity, lipid binding, and allergenic properties. BMC Plant Biol. 2016;16:107. doi: 10.1186/s12870-016-0792-6. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

