A Combined Metabolomic and Metagenomic Approach to Discriminate Raw Milk for the Production of Hard Cheese
- PMID: 33419189
- PMCID: PMC7825538
- DOI: 10.3390/foods10010109
A Combined Metabolomic and Metagenomic Approach to Discriminate Raw Milk for the Production of Hard Cheese
Abstract
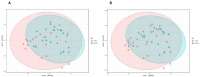
The chemical composition of milk can be significantly affected by different factors across the dairy supply chain, including primary production practices. Among the latter, the feeding system could drive the nutritional value and technological properties of milk and dairy products. Therefore, in this work, a combined foodomics approach based on both untargeted metabolomics and metagenomics was used to shed light onto the impact of feeding systems (i.e., hay vs. a mixed ration based on hay and fresh forage) on the chemical profile of raw milk for the production of hard cheese. In particular, ultra-high-performance liquid chromatography quadrupole time-of-flight mass spectrometry (UHPLC-QTOF) was used to investigate the chemical profile of raw milk (n = 46) collected from dairy herds located in the Po River Valley (Italy) and considering different feeding systems. Overall, a total of 3320 molecular features were putatively annotated across samples, corresponding to 734 unique compound structures, with significant differences (p < 0.05) between the two feeding regimens under investigation. Additionally, supervised multivariate statistics following metabolomics-based analysis allowed us to clearly discriminate raw milk samples according to the feeding systems, also extrapolating the most discriminant metabolites. Interestingly, 10 compounds were able to strongly explain the differences as imposed by the addition of forage in the cows' diet, being mainly glycerophospholipids (i.e., lysophosphatidylethanolamines, lysophosphatidylcholines, and phosphatidylcholines), followed by 5-(3',4'-Dihydroxyphenyl)-gamma-valerolactone-4'-O-glucuronide, 5a-androstan-3a,17b-diol disulfuric acid, and N-stearoyl glycine. The markers identified included both feed-derived (such as phenolic metabolites) and animal-derived compounds (such as lipids and derivatives). Finally, although characterized by a lower prediction ability, the metagenomic profile was found to be significantly correlated to some milk metabolites, with Staphylococcaceae, Pseudomonadaceae, and Dermabacteraceae establishing a higher number of significant correlations with the discriminant metabolites. Therefore, taken together, our preliminary results provide a comprehensive foodomic picture of raw milk samples from different feeding regimens, thus supporting further ad hoc studies investigating the metabolomic and metagenomic changes of milk in all processing conditions.
Keywords: feeding systems; foodomics; lipids; milk quality; secondary metabolites.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Salzano A., Manganiello G., Neglia G., Vinale F., De Nicola D., D’Occhio M., Campanile G. A Preliminary Study on Metabolome Profiles of Buffalo Milk and Corresponding Mozzarella Cheese: Safeguarding the Authenticity and Traceability of Protected Status Buffalo Dairy Products. Molecules. 2020;25:304. doi: 10.3390/molecules25020304. - DOI - PMC - PubMed
-
- Scano P., Carta P., Ibba I., Manis C., Caboni P. An Untargeted Metabolomic Comparison of Milk Composition from Sheep Kept Under Different Grazing Systems. Dairy. 2020;1:30–41. doi: 10.3390/dairy1010004. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

