Upstream Regulator Analysis of Wooden Breast Myopathy Proteomics in Commercial Broilers and Comparison to Feed Efficiency Proteomics in Pedigree Male Broilers
- PMID: 33419207
- PMCID: PMC7825620
- DOI: 10.3390/foods10010104
Upstream Regulator Analysis of Wooden Breast Myopathy Proteomics in Commercial Broilers and Comparison to Feed Efficiency Proteomics in Pedigree Male Broilers
Abstract
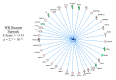
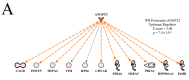
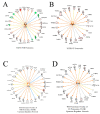
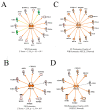
In an effort to understand the apparent trade-off between the continual push for growth performance and the recent emergence of muscle pathologies, shotgun proteomics was conducted on breast muscle obtained at ~8 weeks from commercial broilers with wooden breast (WB) myopathy and compared with that in pedigree male (PedM) broilers exhibiting high feed efficiency (FE). Comparison of the two proteomic datasets was facilitated using the overlay function of Ingenuity Pathway Analysis (IPA) (Qiagen, CA, USA). We focused on upstream regulator analysis and disease-function analysis that provides predictions of activation or inhibition of molecules based on (a) expression of downstream target molecules, (b) the IPA scientific citation database. Angiopoeitin 2 (ANGPT2) exhibited the highest predicted activation Z-score of all molecules in the WB dataset, suggesting that the proteomic landscape of WB myopathy would promote vascularization. Overlaying the FE proteomics data on the WB ANGPT2 upstream regulator network presented no commonality of protein expression and no prediction of ANGPT2 activation. Peroxisome proliferator coactivator 1 alpha (PGC1α) was predicted to be inhibited, suggesting that mitochondrial biogenesis was suppressed in WB. PGC1α was predicted to be activated in high FE pedigree male broilers. Whereas RICTOR (rapamycin independent companion of mammalian target of rapamycin) was predicted to be inhibited in both WB and FE datasets, the predictions were based on different downstream molecules. Other transcription factors predicted to be activated in WB muscle included epidermal growth factor (EGFR), X box binding protein (XBP1), transforming growth factor beta 1 (TGFB1) and nuclear factor (erythroid-derived 2)-like 2 (NFE2L2). Inhibitions of aryl hydrocarbon receptor (AHR), AHR nuclear translocator (ARNT) and estrogen related receptor gamma (ESRRG) were also predicted in the WB muscle. These findings indicate that there are considerable differences in upstream regulators based on downstream protein expression observed in WB myopathy and in high FE PedM broilers that may provide additional insight into the etiology of WB myopathy.
Keywords: feed efficiency; myopathy; proteomics; upstream regulator analysis; wooden breast.
Conflict of interest statement
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures








References
-
- Kuttappan V.A., Bottje W., Ramnathan R., Hartson S.D., Coon C.N., Kong B.-W., Owens C.M., Vazquez-Añon M., Hargis B.M. Proteomic analysis reveals changes in carbohydrate and protein metabolism associated with broiler breast myopathy. Poult. Sci. 2017;96:2992–2999. doi: 10.3382/ps/pex069. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

