Non-Coding RNA-Driven Regulation of rRNA Biogenesis
- PMID: 33419375
- PMCID: PMC7766524
- DOI: 10.3390/ijms21249738
Non-Coding RNA-Driven Regulation of rRNA Biogenesis
Abstract
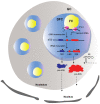
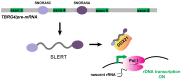
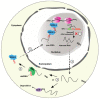
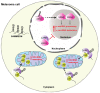
Ribosomal RNA (rRNA) biogenesis takes place in the nucleolus, the most prominent condensate of the eukaryotic nucleus. The proper assembly and integrity of the nucleolus reflects the accurate synthesis and processing of rRNAs which in turn, as major components of ribosomes, ensure the uninterrupted flow of the genetic information during translation. Therefore, the abundant production of rRNAs in a precisely functional nucleolus is of outmost importance for the cell viability and requires the concerted action of essential enzymes, associated factors and epigenetic marks. The coordination and regulation of such an elaborate process depends on not only protein factors, but also on numerous regulatory non-coding RNAs (ncRNAs). Herein, we focus on RNA-mediated mechanisms that control the synthesis, processing and modification of rRNAs in mammals. We highlight the significance of regulatory ncRNAs in rRNA biogenesis and the maintenance of the nucleolar morphology, as well as their role in human diseases and as novel druggable molecular targets.
Keywords: non-coding RNAs; nucleolus; rRNA biogenesis; regulatory RNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

