Performance of Saliva Specimens for the Molecular Detection of SARS-CoV-2 in the Community Setting: Does Sample Collection Method Matter?
- PMID: 33419948
- PMCID: PMC8092755
- DOI: 10.1128/JCM.03033-20
Performance of Saliva Specimens for the Molecular Detection of SARS-CoV-2 in the Community Setting: Does Sample Collection Method Matter?
Abstract
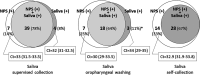
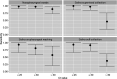
Data on the performance of saliva specimens for diagnosing coronavirus disease 2019 (COVID-19) in ambulatory patients are scarce and inconsistent. We assessed saliva-based specimens for detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by reverse transcriptase PCR (RT-PCR) in the community setting and compared three different collection methods. This prospective study was conducted in three primary care centers. RT-PCR was performed on paired nasopharyngeal swabs (NPS) and saliva samples collected from outpatients with a broad clinical spectrum of illness. To assess differences in collection methods, saliva specimens were obtained in a different way in each of the participating centers: supervised collection (SVC), oropharyngeal washing (OPW), and self-collection (SC). Pairs of NPS and saliva samples from 577 patients (median age, 39 years; 44% men; 42% asymptomatic) were collected and tested, and 120 (20.8%) gave positive results. The overall agreement with NPS results and kappa coefficients (κ) for saliva samples obtained by SVC, OPW, and SC were 95% (κ = 0.85), 93.4% (κ = 0.76), and 93.3% (κ = 0.76), respectively. The sensitivities (95% confidence intervals [95% CI]) of the saliva specimens ranged from 86% (72.6% to 93.7%) for SVC to 66.7% (50.4% to 80%) for SC samples. Sensitivity was higher for samples with lower cycle threshold (CT ) values. The best RT-PCR performance was observed for SVC, with sensitivities (95% CI) of 100% (85.9% to 100%) in symptomatic individuals and 88.9% (50.7% to 99.4%) in asymptomatic individuals at CT values of ≤30. We conclude that saliva is an acceptable specimen for the detection of SARS-CoV-2 in the community setting. Specimens collected under supervision perform comparably to NPS and can effectively identify individuals at higher risk of transmission under real-life conditions.
Keywords: COVID-19; PCR; SARS-CoV-2; diagnostics; saliva.
Copyright © 2021 American Society for Microbiology.
Figures

Similar articles
-
Diagnostic Performance of Self-Collected Saliva Versus Nasopharyngeal Swab for the Molecular Detection of SARS-CoV-2 in the Clinical Setting.Microbiol Spectr. 2021 Dec 22;9(3):e0046821. doi: 10.1128/Spectrum.00468-21. Epub 2021 Nov 3. Microbiol Spectr. 2021. PMID: 34730436 Free PMC article.
-
Saliva as a testing specimen with or without pooling for SARS-CoV-2 detection by multiplex RT-PCR test.PLoS One. 2021 Feb 23;16(2):e0243183. doi: 10.1371/journal.pone.0243183. eCollection 2021. PLoS One. 2021. PMID: 33621263 Free PMC article.
-
Detection of SARS-CoV-2 from Saliva as Compared to Nasopharyngeal Swabs in Outpatients.Viruses. 2020 Nov 17;12(11):1314. doi: 10.3390/v12111314. Viruses. 2020. PMID: 33212817 Free PMC article.
-
Alternative clinical specimens for the detection of SARS-CoV-2: A rapid review.Rev Med Virol. 2021 Jul;31(4):e2185. doi: 10.1002/rmv.2185. Epub 2020 Oct 22. Rev Med Virol. 2021. PMID: 33091200 Review.
-
Diagnosis of SARS-Cov-2 Infection by RT-PCR Using Specimens Other Than Naso- and Oropharyngeal Swabs: A Systematic Review and Meta-Analysis.Diagnostics (Basel). 2021 Feb 21;11(2):363. doi: 10.3390/diagnostics11020363. Diagnostics (Basel). 2021. PMID: 33670020 Free PMC article. Review.
Cited by
-
Comparative evaluation of saliva and nasopharyngeal swab for SARS-CoV-2 detection using RT-qPCR among COVID-19 suspected patients at Jigjiga, Eastern Ethiopia.PLoS One. 2023 Mar 13;18(3):e0282976. doi: 10.1371/journal.pone.0282976. eCollection 2023. PLoS One. 2023. PMID: 36913377 Free PMC article.
-
Saliva as an alternative specimen to nasopharyngeal swabs for COVID-19 diagnosis: Review.Access Microbiol. 2022 May 20;4(5):acmi000366. doi: 10.1099/acmi.0.000366. eCollection 2022 Aug. Access Microbiol. 2022. PMID: 36003360 Free PMC article. Review.
-
Accuracy of saliva and nasopharyngeal sampling for detection of SARS-CoV-2 in community screening: a multicentric cohort study.Eur J Clin Microbiol Infect Dis. 2021 Nov;40(11):2379-2388. doi: 10.1007/s10096-021-04327-x. Epub 2021 Aug 3. Eur J Clin Microbiol Infect Dis. 2021. PMID: 34342768 Free PMC article.
-
Can periodontal pockets and caries lesions act as reservoirs for coronavirus?Mol Oral Microbiol. 2022 Apr;37(2):77-80. doi: 10.1111/omi.12362. Epub 2022 Feb 4. Mol Oral Microbiol. 2022. PMID: 35060684 Free PMC article.
-
Recent advances in RNA sample preparation techniques for the detection of SARS-CoV-2 in saliva and gargle.Trends Analyt Chem. 2023 Aug;165:117107. doi: 10.1016/j.trac.2023.117107. Epub 2023 May 23. Trends Analyt Chem. 2023. PMID: 37317683 Free PMC article. Review.
References
-
- To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, Lau DP, Choi CY, Chen LL, Chan WM, Chan KH, Ip JD, Ng AC, Poon RW, Luo CT, Cheng VC, Chan JF, Hung IF, Chen Z, Chen H, Yuen KY. 2020. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 20:565–574. doi:10.1016/S1473-3099(20)30196-1. - DOI - PMC - PubMed
-
- To KK, Tsang OT, Yip CC, Chan KH, Wu TC, Chan JM, Leung WS, Chik TS, Choi CY, Kandamby DH, Lung DC, Tam AR, Poon RW, Fung AY, Hung IF, Cheng VC, Chan JF, Yuen KY. 2020. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis 71:841–843. doi:10.1093/cid/ciaa149. - DOI - PMC - PubMed
-
- Hanson KE, Barker AP, Hillyard DR, Gilmore N, Barrett JW, Orlandi RR, Shakir SM. 2020. Self-collected anterior nasal and saliva specimens versus health care worker-collected nasopharyngeal swabs for the molecular detection of SARS-CoV-2. J Clin Microbiol 58:e01824-20. doi:10.1128/JCM.01824-20. - DOI - PMC - PubMed
-
- Kojima N, Turner F, Slepnev V, Bacelar A, Deming L, Kodeboyina S, Klausner JD. 2020. Self-collected oral fluid and nasal swab specimens demonstrate comparable sensitivity to clinician-collected nasopharyngeal swab specimens for the detection of SARS-CoV-2. Clin Infect Dis doi:10.1093/cid/ciaa1589. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

