Structures of human dual oxidase 1 complex in low-calcium and high-calcium states
- PMID: 33420071
- PMCID: PMC7794343
- DOI: 10.1038/s41467-020-20466-9
Structures of human dual oxidase 1 complex in low-calcium and high-calcium states
Abstract
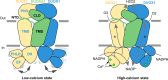
Dual oxidases (DUOXs) produce hydrogen peroxide by transferring electrons from intracellular NADPH to extracellular oxygen. They are involved in many crucial biological processes and human diseases, especially in thyroid diseases. DUOXs are protein complexes co-assembled from the catalytic DUOX subunits and the auxiliary DUOXA subunits and their activities are regulated by intracellular calcium concentrations. Here, we report the cryo-EM structures of human DUOX1-DUOXA1 complex in both high-calcium and low-calcium states. These structures reveal the DUOX1 complex is a symmetric 2:2 hetero-tetramer stabilized by extensive inter-subunit interactions. Substrate NADPH and cofactor FAD are sandwiched between transmembrane domain and the cytosolic dehydrogenase domain of DUOX. In the presence of calcium ions, intracellular EF-hand modules might enhance the catalytic activity of DUOX by stabilizing the dehydrogenase domain in a conformation that allows electron transfer.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

