Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma
- PMID: 33420074
- PMCID: PMC7794332
- DOI: 10.1038/s41467-020-20422-7
Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma
Abstract
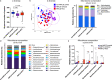
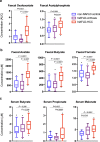
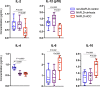
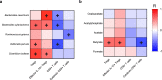
The gut microbiota is reported to modulate the immune response in hepatocellular carcinoma (HCC). Here, we employ metagenomic and metabolomic studies to characterise gut microbiota in patients with non-alcoholic fatty liver disease (NAFLD) related cirrhosis, with or without HCC, and evaluate its effect on the peripheral immune response in an ex vivo model. We find that dysbiosis characterises the microbiota of patients with NAFLD-cirrhosis, with compositional and functional shifts occurring with HCC development. Gene function of the microbiota in NAFLD-HCC supports short chain fatty acid production, and this is confirmed by metabolomic studies. Ex vivo studies show that bacterial extracts from the NAFLD-HCC microbiota, but not from the control groups, elicit a T cell immunosuppressive phenotype, characterised by expansion of regulatory T cells and attenuation of CD8 + T cells. Our study suggest that the gut microbiota in NAFLD-HCC is characterised by a distinctive microbiome/metabolomic profile, and can modulate the peripheral immune response.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

