Visualising G-quadruplex DNA dynamics in live cells by fluorescence lifetime imaging microscopy
- PMID: 33420085
- PMCID: PMC7794231
- DOI: 10.1038/s41467-020-20414-7
Visualising G-quadruplex DNA dynamics in live cells by fluorescence lifetime imaging microscopy
Abstract
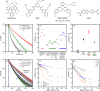
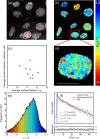
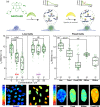
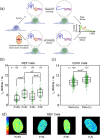
Guanine rich regions of oligonucleotides fold into quadruple-stranded structures called G-quadruplexes (G4s). Increasing evidence suggests that these G4 structures form in vivo and play a crucial role in cellular processes. However, their direct observation in live cells remains a challenge. Here we demonstrate that a fluorescent probe (DAOTA-M2) in conjunction with fluorescence lifetime imaging microscopy (FLIM) can identify G4s within nuclei of live and fixed cells. We present a FLIM-based cellular assay to study the interaction of non-fluorescent small molecules with G4s and apply it to a wide range of drug candidates. We also demonstrate that DAOTA-M2 can be used to study G4 stability in live cells. Reduction of FancJ and RTEL1 expression in mammalian cells increases the DAOTA-M2 lifetime and therefore suggests an increased number of G4s in these cells, implying that FancJ and RTEL1 play a role in resolving G4 structures in cellulo.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Folding and Unfolding of Exogenous G-Rich Oligonucleotides in Live Cells by Fluorescence Lifetime Imaging Microscopy of o-BMVC Fluorescent Probe.Molecules. 2021 Dec 27;27(1):140. doi: 10.3390/molecules27010140. Molecules. 2021. PMID: 35011378 Free PMC article.
-
Imaging G-Quadruplex Nucleic Acids in Live Cells Using Thioflavin T and Fluorescence Lifetime Imaging Microscopy.Anal Chem. 2024 Dec 24;96(51):20223-20229. doi: 10.1021/acs.analchem.4c04207. Epub 2024 Dec 11. Anal Chem. 2024. PMID: 39660854 Free PMC article.
-
Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability.J Biol Chem. 2013 Sep 27;288(39):28217-29. doi: 10.1074/jbc.M113.496463. Epub 2013 Aug 9. J Biol Chem. 2013. PMID: 23935105 Free PMC article.
-
G-quadruplexes and helicases.Nucleic Acids Res. 2016 Mar 18;44(5):1989-2006. doi: 10.1093/nar/gkw079. Epub 2016 Feb 15. Nucleic Acids Res. 2016. PMID: 26883636 Free PMC article. Review.
-
Helicases FANCJ, RTEL1 and BLM Act on Guanine Quadruplex DNA in Vivo.Genes (Basel). 2019 Oct 31;10(11):870. doi: 10.3390/genes10110870. Genes (Basel). 2019. PMID: 31683575 Free PMC article. Review.
Cited by
-
The Cellular Functions and Molecular Mechanisms of G-Quadruplex Unwinding Helicases in Humans.Front Mol Biosci. 2021 Nov 29;8:783889. doi: 10.3389/fmolb.2021.783889. eCollection 2021. Front Mol Biosci. 2021. PMID: 34912850 Free PMC article. Review.
-
New (Iso)quinolinyl-pyridine-2,6-dicarboxamide G-Quadruplex Stabilizers. A Structure-Activity Relationship Study.Pharmaceuticals (Basel). 2021 Jul 13;14(7):669. doi: 10.3390/ph14070669. Pharmaceuticals (Basel). 2021. PMID: 34358095 Free PMC article.
-
Cell redistribution of G quadruplex-structured DNA is associated with morphological changes of nuclei and nucleoli in neurons during tau pathology progression.Brain Pathol. 2025 Mar;35(2):e13262. doi: 10.1111/bpa.13262. Epub 2024 Apr 22. Brain Pathol. 2025. PMID: 38649330 Free PMC article.
-
Optical Probes for Cellular Imaging of G-quadruplexes: Beyond Fluorescence Intensity Probes.Angew Chem Int Ed Engl. 2025 May 26;64(22):e202424931. doi: 10.1002/anie.202424931. Epub 2025 Apr 21. Angew Chem Int Ed Engl. 2025. PMID: 40192564 Free PMC article. Review.
-
Visualization of G-Quadruplexes, i-Motifs and Their Associates.Acta Naturae. 2022 Jul-Sep;14(3):4-18. doi: 10.32607/actanaturae.11705. Acta Naturae. 2022. PMID: 36348720 Free PMC article.
References
-
- Neidle S. Quadruplex nucleic acids as targets for anticancer therapeutics. Nat. Rev. Chem. 2017;1:10. doi: 10.1038/s41570-017-0041. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

