Extracellular vesicle-associated repetitive element DNAs as candidate osteosarcoma biomarkers
- PMID: 33420117
- PMCID: PMC7794510
- DOI: 10.1038/s41598-020-77398-z
Extracellular vesicle-associated repetitive element DNAs as candidate osteosarcoma biomarkers
Abstract
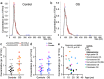
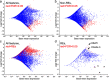
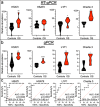
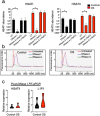
Osteosarcoma (OS) is the most common malignant bone tumor in children and young adults. Despite that high-risk factors have been identified, no test for early detection is available. This study aimed to identify circulating nucleic acid sequences associated with serum extracellular vesicle (EV) preparations at the time of OS diagnosis, as a step towards an OS early detection assay. Sequencing of small nucleic acids extracted from serum EV preparations revealed increased representation of diverse repetitive element sequences in OS patient versus control sera. Analysis of a validation cohort using qPCR of PEG-precipitated EV preparations revealed the over-representation of HSATI, HSATII, LINE1-P1, and Charlie 3 at the DNA but not RNA level, with receiver operating characteristic (ROC) area under the curve (AUC) ≥ 0.90. HSATI and HSATII DNAs co-purified with EVs prepared by precipitation and size exclusion chromatography but not by exosome immunocapture, indicative of packaging in a non-exosomal complex. The consistent over-representation of EV-associated repetitive element DNA sequences suggests their potential utility as biomarkers for OS and perhaps other cancers.
Conflict of interest statement
D.C. and L.C. are inventors on a U.S. provisional patent, Children’s Hospital Los Angeles as applicant, relating to REPETITIVE ELEMENT DNAS.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

