Elucidating the network features and evolutionary attributes of intra- and interspecific protein-protein interactions between human and pathogenic bacteria
- PMID: 33420198
- PMCID: PMC7794237
- DOI: 10.1038/s41598-020-80549-x
Elucidating the network features and evolutionary attributes of intra- and interspecific protein-protein interactions between human and pathogenic bacteria
Abstract
Host-pathogen interaction is one of the most powerful determinants involved in coevolutionary processes covering a broad range of biological phenomena at molecular, cellular, organismal and/or population level. The present study explored host-pathogen interaction from the perspective of human-bacteria protein-protein interaction based on large-scale interspecific and intraspecific interactome data for human and three pathogenic bacterial species, Bacillus anthracis, Francisella tularensis and Yersinia pestis. The network features revealed a preferential enrichment of intraspecific hubs and bottlenecks for both human and bacterial pathogens in the interspecific human-bacteria interaction. Analyses unveiled that these bacterial pathogens interact mostly with human party-hubs that may enable them to affect desired functional modules, leading to pathogenesis. Structural features of pathogen-interacting human proteins indicated an abundance of protein domains, providing opportunities for interspecific domain-domain interactions. Moreover, these interactions do not always occur with high-affinity, as we observed that bacteria-interacting human proteins are rich in protein-disorder content, which correlates positively with the number of interacting pathogen proteins, facilitating low-affinity interspecific interactions. Furthermore, functional analyses of pathogen-interacting human proteins revealed an enrichment in regulation of processes like metabolism, immune system, cellular localization and transport apart from divulging functional competence to bind enzyme/protein, nucleic acids and cell adhesion molecules, necessary for host-microbial cross-talk.
Conflict of interest statement
The authors declare no competing interests.
Figures
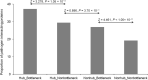
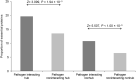



References
-
- Saha S, Sengupta K, Chatterjee P, Basu S, Nasipuri M. Analysis of protein targets in pathogen–host interaction in infectious diseases: A case study on Plasmodium falciparum and Homo sapiens interaction network. Brief. Funct. Genom. 2017;17:441–450. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

