False negatives in GBA1 sequencing due to polymerase dependent allelic imbalance
- PMID: 33420335
- PMCID: PMC7794395
- DOI: 10.1038/s41598-020-80564-y
False negatives in GBA1 sequencing due to polymerase dependent allelic imbalance
Abstract
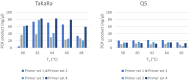
A variant in the GBA1 gene is one of the most common genetic risk factors to develop Parkinson's disease (PD). Here the serendipitous finding is reported of a polymerase dependent allelic imbalance when using next generation sequencing, potentially resulting in false-negative results when the allele frequency falls below the variant calling threshold (by default commonly at 30%). The full GBA1 gene was sequenced using next generation sequencing on saliva derived DNA from PD patients. Four polymerase chain reaction conditions were varied in twelve samples, to investigate the effect on allelic imbalance: (1) the primers (n = 4); (2) the polymerase enzymes (n = 2); (3) the primer annealing temperature (Ta) specified for the used polymerase; and (4) the amount of DNA input. Initially, 1295 samples were sequenced using Q5 High-Fidelity DNA Polymerase. 112 samples (8.6%) had an exonic variant and an additional 104 samples (8.0%) had an exonic variant that did not pass the variant frequency calling threshold of 30%. After changing the polymerase to TaKaRa LA Taq DNA Polymerase Hot-Start Version: RR042B, all samples had an allele frequency passing the calling threshold. Allele frequency was unaffected by a change in primer, annealing temperature or amount of DNA input. Sequencing of the GBA1 gene using next generation sequencing might be susceptible to a polymerase specific allelic imbalance, which can result in a large amount of flase-negative results. This was resolved in our case by changing the polymerase. Regions displaying low variant calling frequencies in GBA1 sequencing output in previous and future studies might warrant additional scrutiny.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

