PRMT1 enhances oncogenic arginine methylation of NONO in colorectal cancer
- PMID: 33420374
- PMCID: PMC7892343
- DOI: 10.1038/s41388-020-01617-0
PRMT1 enhances oncogenic arginine methylation of NONO in colorectal cancer
Abstract
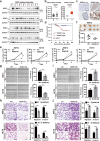
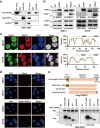
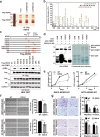
Arginine methylation is an important posttranslational modification catalyzed by protein arginine methyltransferases (PRMTs). However, the role of PRMTs in colorectal cancer (CRC) progression is not well understood. Here we report that non-POU domain-containing octamer-binding protein (NONO) is overexpressed in CRC tissue and is a potential marker for poor prognosis in CRC patients. NONO silencing resulted in decreased proliferation, migration, and invasion of CRC cells, whereas overexpression had the opposite effect. In a xenograft model, tumors derived from NONO-deficient CRC cells were smaller than those derived from wild-type (WT) cells, and PRMT1 inhibition blocked CRC xenograft progression. A mass spectrometry analysis indicated that NONO is a substrate of PRMT1. R251 of NONO was asymmetrically dimethylated by PRMT1 in vitro and in vivo. Compared to NONO WT cells, NONO R251K mutant-expressing CRC cells showed reduced proliferation, migration, and invasion, and PRMT1 knockdown or pharmacological inhibition abrogated the malignant phenotype associated with NONO asymmetric dimethylation in both KRAS WT and mutant CRC cells. Compared to adjacent normal tissue, PRMT1 was highly expressed in the CRC zone in clinical specimens, which was correlated with poor overall survival in patients with locally advanced CRC. These results demonstrate that PRMT1-mediated methylation of NONO at R251 promotes CRC growth and metastasis, and suggest that PRMT1 inhibition may be an effective therapeutic strategy for CRC treatment regardless of KRAS mutation status.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Sauer R, Liersch T, Merkel S, Fietkau R, Hohenberger W, Hess C, et al. Preoperative versus postoperative chemoradiotherapy for locally advanced rectal cancer: results of the German CAO/ARO/AIO-94 randomized phase III trial after a median follow-up of 11 years. J Clin Oncol. 2012;30:1926–33. - PubMed
-
- Dienstmann R, Salazar R, Tabernero J. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J Clin Oncol. 2015;33:1787–96. - PubMed
-
- Lièvre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–9. - PubMed
-
- Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N. Engl J Med. 2009;360:1408–17. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

