Combined whole cell wall analysis and streamlined in silico carbohydrate-active enzyme discovery to improve biocatalytic conversion of agricultural crop residues
- PMID: 33422151
- PMCID: PMC7797155
- DOI: 10.1186/s13068-020-01869-8
Combined whole cell wall analysis and streamlined in silico carbohydrate-active enzyme discovery to improve biocatalytic conversion of agricultural crop residues
Erratum in
-
Correction to: Combined whole cell wall analysis and streamlined in silico carbohydrate‑active enzyme discovery to improve biocatalytic conversion of agricultural crop residues.Biotechnol Biofuels. 2021 Feb 8;14(1):40. doi: 10.1186/s13068-021-01888-z. Biotechnol Biofuels. 2021. PMID: 33557907 Free PMC article. No abstract available.
Abstract
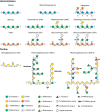
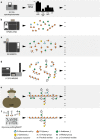
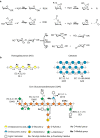
The production of biofuels as an efficient source of renewable energy has received considerable attention due to increasing energy demands and regulatory incentives to reduce greenhouse gas emissions. Second-generation biofuel feedstocks, including agricultural crop residues generated on-farm during annual harvests, are abundant, inexpensive, and sustainable. Unlike first-generation feedstocks, which are enriched in easily fermentable carbohydrates, crop residue cell walls are highly resistant to saccharification, fermentation, and valorization. Crop residues contain recalcitrant polysaccharides, including cellulose, hemicelluloses, pectins, and lignin and lignin-carbohydrate complexes. In addition, their cell walls can vary in linkage structure and monosaccharide composition between plant sources. Characterization of total cell wall structure, including high-resolution analyses of saccharide composition, linkage, and complex structures using chromatography-based methods, nuclear magnetic resonance, -omics, and antibody glycome profiling, provides critical insight into the fine chemistry of feedstock cell walls. Furthermore, improving both the catalytic potential of microbial communities that populate biodigester reactors and the efficiency of pre-treatments used in bioethanol production may improve bioconversion rates and yields. Toward this end, knowledge and characterization of carbohydrate-active enzymes (CAZymes) involved in dynamic biomass deconstruction is pivotal. Here we overview the use of common "-omics"-based methods for the study of lignocellulose-metabolizing communities and microorganisms, as well as methods for annotation and discovery of CAZymes, and accurate prediction of CAZyme function. Emerging approaches for analysis of large datasets, including metagenome-assembled genomes, are also discussed. Using complementary glycomic and meta-omic methods to characterize agricultural residues and the microbial communities that digest them provides promising streams of research to maximize value and energy extraction from crop waste streams.
Keywords: Agriculture; Biomass conversion; Carbohydrate-active enzyme; Crop residues; Functional genomics; Glycosidic linkage analysis; Phylogeny; Plant cell wall.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Callegari A, Bolognesi S, Cecconet D, Capodaglio AG. Production technologies, current role, and future prospects of biofuels feedstocks: a state-of-the-art review. Crit Rev Environ Sci Technol. 2020;50(4):384–436. doi: 10.1080/10643389.2019.1629801. - DOI
-
- Campanaro S, Treu L, Kougias PG, De Francisci D, Valle G, Angelidaki I. Metagenomic analysis and functional characterization of the biogas microbiome using high throughput shotgun sequencing and a novel binning strategy. Biotechnol Biofuels. 2016;9(1):26. doi: 10.1186/s13068-016-0441-1. - DOI - PMC - PubMed
-
- Tomei J, Helliwell R. Food versus fuel? Going beyond biofuels. Land Use Policy. 2016;56:320–326. doi: 10.1016/j.landusepol.2015.11.015. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

