ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation
- PMID: 33422265
- PMCID: PMC7832235
- DOI: 10.1016/j.devcel.2020.12.010
ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation
Abstract
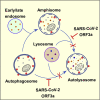
Autophagy acts as a cellular surveillance mechanism to combat invading pathogens. Viruses have evolved various strategies to block autophagy and even subvert it for their replication and release. Here, we demonstrated that ORF3a of the COVID-19 virus SARS-CoV-2 inhibits autophagy activity by blocking fusion of autophagosomes/amphisomes with lysosomes. The late endosome-localized ORF3a directly interacts with and sequestrates the homotypic fusion and protein sorting (HOPS) component VPS39, thereby preventing HOPS complex from interacting with the autophagosomal SNARE protein STX17. This blocks assembly of the STX17-SNAP29-VAMP8 SNARE complex, which mediates autophagosome/amphisome fusion with lysosomes. Expression of ORF3a also damages lysosomes and impairs their function. SARS-CoV-2 virus infection blocks autophagy, resulting in accumulation of autophagosomes/amphisomes, and causes late endosomal sequestration of VPS39. Surprisingly, ORF3a from the SARS virus SARS-CoV fails to interact with HOPS or block autophagy. Our study reveals a mechanism by which SARS-CoV-2 evades lysosomal destruction and provides insights for developing new strategies to treat COVID-19.
Keywords: COVID-19; DMV; HOPS; ORF3a; SARS-CoV-2; SNARE; autophagy.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Comment in
-
Autophagosome maturation stymied by SARS-CoV-2.Dev Cell. 2021 Feb 22;56(4):400-402. doi: 10.1016/j.devcel.2021.02.002. Dev Cell. 2021. PMID: 33621488 Free PMC article.
Similar articles
-
Myelin basic protein antagonizes the SARS-CoV-2 protein ORF3a-induced autophagy inhibition.Biochimie. 2024 Oct;225:1-9. doi: 10.1016/j.biochi.2024.04.011. Epub 2024 May 3. Biochimie. 2024. PMID: 38703943
-
Regulation of autophagy by SARS-CoV-2: The multifunctional contributions of ORF3a.J Med Virol. 2023 Jul;95(7):e28959. doi: 10.1002/jmv.28959. J Med Virol. 2023. PMID: 37485696 Review.
-
ORF3a of SARS-CoV-2 promotes lysosomal exocytosis-mediated viral egress.Dev Cell. 2021 Dec 6;56(23):3250-3263.e5. doi: 10.1016/j.devcel.2021.10.006. Epub 2021 Oct 11. Dev Cell. 2021. PMID: 34706264 Free PMC article.
-
Impairment of autophagosome-lysosome fusion in the buff mutant mice with the VPS33A(D251E) mutation.Autophagy. 2015;11(9):1608-22. doi: 10.1080/15548627.2015.1072669. Autophagy. 2015. PMID: 26259518 Free PMC article.
-
Evolutionarily conserved role and physiological relevance of a STX17/Syx17 (syntaxin 17)-containing SNARE complex in autophagosome fusion with endosomes and lysosomes.Autophagy. 2013 Oct;9(10):1642-6. doi: 10.4161/auto.25684. Epub 2013 Jul 22. Autophagy. 2013. PMID: 24113031 Review.
Cited by
-
Post-COVID-19 Parkinsonism and Parkinson's Disease Pathogenesis: The Exosomal Cargo Hypothesis.Int J Mol Sci. 2022 Aug 28;23(17):9739. doi: 10.3390/ijms23179739. Int J Mol Sci. 2022. PMID: 36077138 Free PMC article. Review.
-
Lipid compartments and lipid metabolism as therapeutic targets against coronavirus.Front Immunol. 2023 Dec 1;14:1268854. doi: 10.3389/fimmu.2023.1268854. eCollection 2023. Front Immunol. 2023. PMID: 38106410 Free PMC article. Review.
-
Extracellular vesicle proteomics and phosphoproteomics identify pathways for increased risk in patients hospitalized with COVID-19 and type 2 diabetes mellitus.Diabetes Res Clin Pract. 2023 Mar;197:110565. doi: 10.1016/j.diabres.2023.110565. Epub 2023 Feb 1. Diabetes Res Clin Pract. 2023. PMID: 36736734 Free PMC article.
-
Friend or Foe? Implication of the autophagy-lysosome pathway in SARS-CoV-2 infection and COVID-19.Int J Biol Sci. 2022 Jul 11;18(12):4690-4703. doi: 10.7150/ijbs.72544. eCollection 2022. Int J Biol Sci. 2022. PMID: 35874956 Free PMC article. Review.
-
A bibliometric analysis of programmed cell death in acute lung injury/acute respiratory distress syndrome from 2000 to 2022.Heliyon. 2023 Sep 1;9(9):e19759. doi: 10.1016/j.heliyon.2023.e19759. eCollection 2023 Sep. Heliyon. 2023. PMID: 37809536 Free PMC article.
References
-
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. - PMC - PubMed
-
- Balderhaar H.J., Ungermann C. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J. Cell Sci. 2013;126:1307–1316. - PubMed
-
- Bonifacino J.S., Rojas R. Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 2006;7:568–579. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

