Off-the-Shelf, Immune-Compatible Human Embryonic Stem Cells Generated Via CRISPR-Mediated Genome Editing
- PMID: 33423156
- PMCID: PMC8166669
- DOI: 10.1007/s12015-020-10113-7
Off-the-Shelf, Immune-Compatible Human Embryonic Stem Cells Generated Via CRISPR-Mediated Genome Editing
Abstract
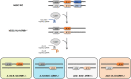
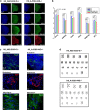
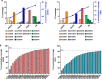
Human embryonic stem cells (hESCs) hold promise in regenerative medicine but allogeneic immune rejections caused by highly polymorphic human leukocyte antigens (HLAs) remain a barrier to their clinical applications. Here, we used a CRISPR/Cas9-mediated HLA-editing strategy to generate a variety of HLA homozygous-like hESC lines from pre-established hESC lines. We edited four pre-established HLA-heterozygous hESC lines and created a mini library of 14 HLA-edited hESC lines in which single HLA-A and HLA-B alleles and both HLA-DR alleles are disrupted. The HLA-edited hESC derivatives elicited both low T cell- and low NK cell-mediated immune responses. Our library would cover about 40% of the Asian-Pacific population. We estimate that HLA-editing of only 19 pre-established hESC lines would give rise to 46 different hESC lines to cover 90% of the Asian-Pacific population. This study offers an opportunity to generate an off-the-shelf HLA-compatible hESC bank, available for immune-compatible cell transplantation, without embryo destruction. Graphical Abstract.
Keywords: CRISPR/Cas9; HLA homozygous-like hESCs; HLA-editing; Human embryonic stem cells; Immune-compatible hESC banking.
Conflict of interest statement
J.-S.K. is a cofounder of, and holds stock in, ToolGen, Inc.
Figures



References
-
- Bradley JA, Bolton EM, Pedersen RA. Stem cell medicine encounters the immune system. Nature Reviews Immunology. 2002;2(11):859–871. - PubMed
-
- Edgerly CH, Weimer ET. The past, present, and future of HLA typing in transplantation. Methods in Molecular Biology. 2018;1802:1–10. - PubMed
-
- Drukker M, Katchman H, Katz G, Even-Tov Friedman S, Shezen E, Hornstein E, Mandelboim O, Reisner Y, Benvenisty N. Human embryonic stem cells and their differentiated derivatives are less susceptible to immune rejection than adult cells. Stem Cells. 2006;24(2):221–229. - PubMed
-
- Li L, Baroja ML, Majumdar A, Chadwick K. Human embryonic stem cells possess immune-privileged properties. Stem. 2004;22:448–456. - PubMed
-
- Lin G, Ou-Yang Q, Qian X, Lu G. Construction of human embryonic stem cell banks: Prospects for tissue matching. In: Fairchild PJ, editor. The immunological barriers to regenerative medicine. New York: Springer; 2013. pp. 111–128.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

