Untangling a complex web: Computational analyses of tumor molecular profiles to decode driver mechanisms
- PMID: 33423960
- PMCID: PMC7902422
- DOI: 10.1016/j.jgg.2020.11.001
Untangling a complex web: Computational analyses of tumor molecular profiles to decode driver mechanisms
Abstract
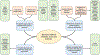
Genome-scale studies focusing on molecular profiling of cancers across tissue types have revealed a plethora of aberrations across the genomic, transcriptomic, and epigenomic scales. The significant molecular heterogeneity across individual tumors even within the same tissue context complicates decoding the key etiologic mechanisms of this disease. Furthermore, it is increasingly likely that biologic mechanisms underlying the pathobiology of cancer involve multiple molecular entities interacting across functional scales. This has motivated the development of computational approaches that integrate molecular measurements with prior biological knowledge in increasingly intricate ways to enable the discovery of driver genomic aberrations across cancers. Here, we review diverse methodological approaches that have powered significant advances in our understanding of the genomic underpinnings of cancer at the cohort and at the individual tumor scales. We outline the key advances and challenges in the computational discovery of cancer mechanisms while motivating the development of systems biology approaches to comprehensively decode the biologic drivers of this complex disease.
Keywords: Functional impact; Multiomics integration; Mutational significance; Mutations; Pan-cancer analysis; Systems biology.
Copyright © 2020 Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, and Genetics Society of China. Published by Elsevier Ltd. All rights reserved.
Conflict of interest statement
Figures



References
-
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinsk M, Jager N, Jones DTW, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt ANJ, Valdes-Mas R, van Buuren MM, van ‘t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR, Genome APC, Consortium IBC, Consortium IM-S, PedBrain I, 2013. Signatures of mutational processes in human cancer. Nature 500, 415–421. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

