Discovery of New Fusion Inhibitor Peptides against SARS-CoV-2 by Targeting the Spike S2 Subunit
- PMID: 33424013
- PMCID: PMC8094075
- DOI: 10.4062/biomolther.2020.201
Discovery of New Fusion Inhibitor Peptides against SARS-CoV-2 by Targeting the Spike S2 Subunit
Abstract
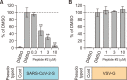
A novel coronavirus, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), caused a worldwide pandemic. Our aim in this study is to produce new fusion inhibitors against SARS-CoV-2, which can be the basis for developing new antiviral drugs. The fusion core comprising the heptad repeat domains (HR1 and HR2) of SARS-CoV-2 spike (S) were used to design the peptides. A total of twelve peptides were generated, comprising a short or truncated 24-mer (peptide #1), a long 36-mer peptide (peptide #2), and ten peptide #2 analogs. In contrast to SARS-CoV, SARS-CoV-2 S-mediated cell-cell fusion cannot be inhibited with a minimal length, 24-mer peptide. Peptide #2 demonstrated potent inhibition of SARS-CoV-2 S-mediated cell-cell fusion at 1 µM concentration. Three peptide #2 analogs showed IC50 values in the low micromolar range (4.7-9.8 µM). Peptide #2 inhibited the SARS-CoV-2 pseudovirus assay at IC50=1.49 µM. Given their potent inhibition of viral activity and safety and lack of cytotoxicity, these peptides provide an attractive avenue for the development of new prophylactic and therapeutic agents against SARS-CoV-2.
Keywords: Antiviral drugs; COVID-19; Fusion inhibitors; SARS-CoV-2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Design of Potent Membrane Fusion Inhibitors against SARS-CoV-2, an Emerging Coronavirus with High Fusogenic Activity.J Virol. 2020 Jul 1;94(14):e00635-20. doi: 10.1128/JVI.00635-20. Print 2020 Jul 1. J Virol. 2020. PMID: 32376627 Free PMC article.
-
Repurposing fusion inhibitor peptide against SARS-CoV-2.J Comput Chem. 2021 Dec 15;42(32):2283-2293. doi: 10.1002/jcc.26758. Epub 2021 Sep 30. J Comput Chem. 2021. PMID: 34591335
-
Discovery of New Potent anti-MERS CoV Fusion Inhibitors.Front Pharmacol. 2021 Jun 2;12:685161. doi: 10.3389/fphar.2021.685161. eCollection 2021. Front Pharmacol. 2021. PMID: 34149429 Free PMC article.
-
Targetable elements in SARS-CoV-2 S2 subunit for the design of pan-coronavirus fusion inhibitors and vaccines.Signal Transduct Target Ther. 2023 May 10;8(1):197. doi: 10.1038/s41392-023-01472-x. Signal Transduct Target Ther. 2023. PMID: 37164987 Free PMC article. Review.
-
MERS-CoV spike protein: a key target for antivirals.Expert Opin Ther Targets. 2017 Feb;21(2):131-143. doi: 10.1080/14728222.2017.1271415. Epub 2016 Dec 21. Expert Opin Ther Targets. 2017. PMID: 27936982 Free PMC article. Review.
Cited by
-
Helix-based screening with structure prediction using artificial intelligence has potential for the rapid development of peptide inhibitors targeting class I viral fusion.RSC Chem Biol. 2023 Nov 7;5(2):131-140. doi: 10.1039/d3cb00166k. eCollection 2024 Feb 7. RSC Chem Biol. 2023. PMID: 38333196 Free PMC article.
-
Molnupiravir-A Novel Oral Anti-SARS-CoV-2 Agent.Antibiotics (Basel). 2021 Oct 23;10(11):1294. doi: 10.3390/antibiotics10111294. Antibiotics (Basel). 2021. PMID: 34827232 Free PMC article.
-
Drug Discovery of New Anti-Inflammatory Compounds by Targeting Cyclooxygenases.Pharmaceuticals (Basel). 2022 Feb 24;15(3):282. doi: 10.3390/ph15030282. Pharmaceuticals (Basel). 2022. PMID: 35337080 Free PMC article.
-
Targeting an evolutionarily conserved "E-L-L" motif in spike protein to identify a small molecule fusion inhibitor against SARS-CoV-2.PNAS Nexus. 2022 Oct 22;1(5):pgac198. doi: 10.1093/pnasnexus/pgac198. eCollection 2022 Nov. PNAS Nexus. 2022. PMID: 36712339 Free PMC article.
-
The Possible Mechanistic Basis of Individual Susceptibility to Spike Protein Injury.Adv Virol. 2025 Jun 24;2025:7990876. doi: 10.1155/av/7990876. eCollection 2025. Adv Virol. 2025. PMID: 40599824 Free PMC article. Review.
References
-
- Bashyal S., Noh G., Keum T., Choi Y. W., Lee S. Cell penetrating peptides as an innovative approach for drug delivery; then, present and the future. J. Pharm. Investig. 2016;46:205–220. doi: 10.1007/s40005-016-0253-0. - DOI
-
- Chu L. H. M., Chan S. H., Tsai S. N., Wang Y., Cheng C. H. K., Wong K. B., Waye M. M. Y., Ngai S. M. Fusion core structure of the severe acute respiratory syndrome coronavirus (SARS-CoV): in search of potent SARS-CoV entry inhibitors. J. Cell. Biochem. 2008;104:2335–2347. doi: 10.1002/jcb.21790. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

