Identification of potential inhibitors of Zika virus NS5 RNA-dependent RNA polymerase through virtual screening and molecular dynamic simulations
- PMID: 33424251
- PMCID: PMC7783101
- DOI: 10.1016/j.jsps.2020.10.005
Identification of potential inhibitors of Zika virus NS5 RNA-dependent RNA polymerase through virtual screening and molecular dynamic simulations
Abstract
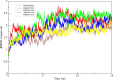
Zika virus (ZIKV) is one of the mosquito borne flavivirus with several outbreaks in past few years in tropical and subtropical regions. The non-structural proteins of flaviviruses are suitable active targets for inhibitory drugs due to their role in pathogenicity. In ZIKV, the non-structural protein 5 (NS5) RNA-Dependent RNA polymerase replicates its genome. Here we have performed virtual screening to identify suitable ligands that can potentially halt the ZIKV NS5 RNA dependent RNA polymerase (RdRp). During this process, we searched and screened a library of ligands against ZIKV NS5 RdRp. The selected ligands with significant binding energy and ligand-receptor interactions were further processed. Among the selected docked conformations, top five was further optimized at atomic level using molecular dynamic simulations followed by binding free energy calculations. The interactions of ligands with the target structure of ZIKV RdRp revealed that they form strong bonds within the active sites of the receptor molecule. The efficacy of these drugs against ZIKV can be further analyzed through in-vitro and in-vivo studies.
Keywords: Binding free energy; Docking studies; Molecular dynamics; NS5 RNA-dependent RNA polymerase; Virtual screening; Zika virus.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Discovery of Bispecific Lead Compounds from Azadirachta indica against ZIKA NS2B-NS3 Protease and NS5 RNA Dependent RNA Polymerase Using Molecular Simulations.Molecules. 2022 Apr 15;27(8):2562. doi: 10.3390/molecules27082562. Molecules. 2022. PMID: 35458761 Free PMC article.
-
Identification of Potent Zika Virus NS5 RNA-Dependent RNA Polymerase Inhibitors Combining Virtual Screening and Biological Assays.Int J Mol Sci. 2023 Jan 18;24(3):1900. doi: 10.3390/ijms24031900. Int J Mol Sci. 2023. PMID: 36768218 Free PMC article.
-
Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase.J Virol. 2020 Oct 14;94(21):e00794-20. doi: 10.1128/JVI.00794-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32796069 Free PMC article.
-
The Potential Role of the ZIKV NS5 Nuclear Spherical-Shell Structures in Cell Type-Specific Host Immune Modulation during ZIKV Infection.Cells. 2019 Nov 26;8(12):1519. doi: 10.3390/cells8121519. Cells. 2019. PMID: 31779251 Free PMC article. Review.
-
The Transactions of NS3 and NS5 in Flaviviral RNA Replication.Adv Exp Med Biol. 2018;1062:147-163. doi: 10.1007/978-981-10-8727-1_11. Adv Exp Med Biol. 2018. PMID: 29845531 Review.
Cited by
-
In silico screening for potential inhibitors from the phytocompounds of Carica papaya against Zika virus NS5 protein.F1000Res. 2024 Jun 25;12:655. doi: 10.12688/f1000research.134956.2. eCollection 2023. F1000Res. 2024. PMID: 39132582 Free PMC article.
-
A review on structural genomics approach applied for drug discovery against three vector-borne viral diseases: Dengue, Chikungunya and Zika.Virus Genes. 2022 Jun;58(3):151-171. doi: 10.1007/s11262-022-01898-5. Epub 2022 Apr 8. Virus Genes. 2022. PMID: 35394596 Review.
-
Chemoinformatic Design and Profiling of Derivatives of Dasabuvir, Efavirenz, and Tipranavir as Potential Inhibitors of Zika Virus RNA-Dependent RNA Polymerase and Methyltransferase.ACS Omega. 2022 Sep 6;7(37):33330-33348. doi: 10.1021/acsomega.2c03945. eCollection 2022 Sep 20. ACS Omega. 2022. PMID: 36157724 Free PMC article.
-
Flavivirus: From Structure to Therapeutics Development.Life (Basel). 2021 Jun 25;11(7):615. doi: 10.3390/life11070615. Life (Basel). 2021. PMID: 34202239 Free PMC article. Review.
-
Deep Generative Models for the Discovery of Antiviral Peptides Targeting Dengue Virus: A Systematic Review.Int J Mol Sci. 2025 Jun 26;26(13):6159. doi: 10.3390/ijms26136159. Int J Mol Sci. 2025. PMID: 40649934 Free PMC article. Review.
References
-
- Abbasi S., Raza S., Azam S.S., Liedl K.R., Fuchs J.E. Interaction mechanisms of a melatonergic inhibitor in the melatonin synthesis pathway. J. Mol. Liq. 2016;221:507–517.
-
- Abro A., Azam S.S. Binding free energy based analysis of arsenic (+ 3 oxidation state) methyltransferase with S-adenosylmethionine. J. Mol. Liq. 2016;220:375–382.
-
- Ahmad N., Badshah S.L., Junaid M., Ur Rehman A., Muhammad A., Khan K. Structural insights into the Zika virus NS1 protein inhibition using a computational approach. J. Biomol. Struct. Dyn. 2020:1–8. - PubMed
-
- Ahmad N., Farman A., Badshah S.L., Rahman A.U., ur Rashid H., Khan K. Molecular modeling, simulation and docking study of ebola virus glycoprotein. J. Mol. Graph. Model. 2017;72:266–271. - PubMed
-
- Ahmad N., Rehman A.U., Badshah S.L., Ullah A., Mohammad A., Khan K. Molecular dynamics simulation of zika virus NS5 RNA dependent RNA polymerase with selected novel non-nucleoside inhibitors. J. Mol. Struct. 2020;1203
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

