Analysis of bacterial communities in sponges and coral inhabiting Red Sea, using barcoded 454 pyrosequencing
- PMID: 33424375
- PMCID: PMC7783839
- DOI: 10.1016/j.sjbs.2020.11.021
Analysis of bacterial communities in sponges and coral inhabiting Red Sea, using barcoded 454 pyrosequencing
Abstract
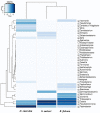
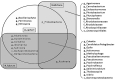
Microbial communities are linked with marine sponge are diverse in their structure and function. Our understanding of the sponge-associated microbial diversity is limited especially from Red Sea in Saudi Arabia where few species of sponges have been studied. Here we used pyrosequencing to study two marine sponges and coral species sampled from Obhur region from Red sea in Jeddah. A total of 168 operational taxonomic units (OTUs) were identified from Haliclona caerulea, Stylissa carteri and Rhytisma fulvum. Taxonomic identification of tag sequences of 16S ribosomal RNA revealed 6 different bacterial phyla and 9 different classes. A proportion of unclassified reads were was also observed in sponges and coral sample. We found diverse bacterial communities associated with two sponges and a coral sample. Diversity and richness estimates based on OUTs revealed that sponge H. caerulea had significantly high bacterial diversity. The identified OTUs showed unique clustering in three sponge samples as revealed by Principal coordinate analysis (PCoA). Proteobacteria (88-95%) was dominant phyla alonwith Bacteroidetes, Planctomycetes, Cyanobacteria, Firmicutes and Nitrospirae. Seventeen different genera were identified where genus Pseudoalteromonas was dominant in all three samples. This is first study to assess bacterial communities of sponge and coral sample that have never been studied before to unravel their microbial communities using 454-pyrosequencing method.
Keywords: 454 pyrosequencing; Bacterial diversity; Marine sponges; Proteobacteria; Red sea.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Bavestrello G., Arillo A., Calcinai B., Cattaneo-Vietti R., Cerrano C., Gaino E., Penna A., Sara M. Parasitic diatoms inside antarctic sponges. Biol. Bull. 2000;198(1):29–33. - PubMed
-
- Bayer K., Kamke J., Hentschel U. Quantification of bacterial and archaeal symbionts in high and low microbial abundance sponges using real-time PCR. FEMS Microbiol. Ecol. 2014;89:679–690. - PubMed
-
- Bell J.J. The functional roles of marine sponges. Estuar. Coast. Shelf Sci. 2008;79(3):341–353.
LinkOut - more resources
Full Text Sources
Other Literature Sources

