In silico evaluation of flavonoids as effective antiviral agents on the spike glycoprotein of SARS-CoV-2
- PMID: 33424398
- PMCID: PMC7783825
- DOI: 10.1016/j.sjbs.2020.11.049
In silico evaluation of flavonoids as effective antiviral agents on the spike glycoprotein of SARS-CoV-2
Abstract
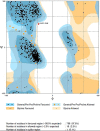
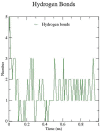
The novel coronavirus pandemic has spread over in 213 countries as of July 2020. Approximately 12 million people have been infected so far according to the reports from World Health Organization (WHO). Preventive measures are being taken globally to avoid the rapid spread of virus. In the current study, an in silico approach is carried out as a means of inhibiting the spike protein of the novel coronavirus by flavonoids from natural sources that possess both antiviral and anti-inflammatory properties. The methodology is focused on molecular docking of 10 flavonoid compounds that are docked with the spike protein of SARS-CoV-2, to determine the highest binding affinity at the binding site. Molecular dynamics simulation was carried out with the flavonoid-protein complex showing the highest binding affinity and highest interactions. The flavonoid naringin showed the least binding energy of -9.8 Kcal/mol with the spike protein which was compared with the standard drug, dexamethasone which is being repurposed to treat critically ill patients. MD simulation was carried out on naringin-spike protein complex for their conformational stability in the active site of the novel coronavirus spike protein. The RMSD of the complex appeared to be more stable when compared to that of the protein from 0.2 nm to 0.4 nm. With the aid of this in silico approach further in vitro studies can be carried out on these flavonoids against the novel coronavirus as a means of viral protein inhibitors.
Keywords: Antiviral; COVID-19, Coronavirus Disease 2019; CoVs, Coronaviruses; Covid-19; Docking; Flavonoids; MD simulations; PDB, Protein Data Bank; RMSD, Root Mean Square Deviation; RMSF, Root Mean Square Fluctuation; Rg, Radius of Gyration; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures









References
-
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindah E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. - DOI
-
- Adhikari S.P., Meng S., Wu Y., Mao Y., Ye R., Wang Q., Sun C., Sylvia S., Rozelle S., Raat H., Zhou H. A literature review of 2019 Novel Coronavirus during the early outbreak period: Epidemiology, causes, clinical manifestation and diagnosis, prevention and control. Infect. Dis. Poverty. 2020;9:1–12. - PMC - PubMed
-
- Ahmadi A., Hassandarvish P., Lani R., Yadollahi P., Jokar A., Bakar S.A., Zandi K. Inhibition of chikungunya virus replication by hesperetin and naringenin. RSC Adv. 2016;6:69421–69430. doi: 10.1039/c6ra16640g. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

