Acinetobacter baumannii as a community foodborne pathogen: Peptide mass fingerprinting analysis, genotypic of biofilm formation and phenotypic pattern of antimicrobial resistance
- PMID: 33424412
- PMCID: PMC7783781
- DOI: 10.1016/j.sjbs.2020.11.052
Acinetobacter baumannii as a community foodborne pathogen: Peptide mass fingerprinting analysis, genotypic of biofilm formation and phenotypic pattern of antimicrobial resistance
Abstract
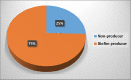
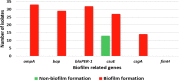
Acinetobacter baumannii (A. baumannii) is one of the most common Gram-negative pathogens that represent a major threat to human life. Because the prevalence of Multidrug-resistant biofilm-forming A. baumannii is increasing all over the world, this may lead to outbreaks of hospital infections. Nonetheless, the role of raw meat as a reservoir for A. baumannii remains unclear. Here our research was aimed to exhibit the frequency, precise identification, and genotyping of biofilm-related genes as well as antimicrobial resistance of A. baumannii isolates of raw meat specimens. Fifty-five A. baumannii strains were recovered from 220 specimens of different animal meat and then identified by Peptide Mass Fingerprinting Technique (PMFT). All identified isolates were genotyped by the qPCR method for the existence of biofilm-related genes (ompA, bap, blaPER-1, csuE, csgA, and fimH). In addition, the antimicrobial resistance against A. baumannii was detected by the Kirby-Bauer method. Based on our findings, the frequency rate of 55 A. baumannii isolates was 46.55%, 32.50%, 15.00%, and 9.68% of sheep, chicken, cow, and camel raw meat samples, respectively. The PMFT was able to identify all strains by 100%. the percentages of csuE, ompA, blaPER-1, bap, and csgA genes in biofilm and non-biofilm producer A. baumannii were 72.73%, 60%, 58.2%, 52.74%, and 25.45%, respectively. In contrast, the fimH was not detected in all non-biofilm and biofilm producer strains. The ompA, bap, blaPER-1, csgA were detected only in biofilm-producing A. baumannii isolates. The maximum degree of resistance was observed against amoxicillin/clavulanic acid (89.10%), gentamicin (74.55%), tetracycline (72.73%), ampicillin (65.45%), and tobramycin (52.73%). In conclusion, our investigation demonstrated the high incidence of multi-drug resistant A. baumannii in raw meat samples, with a high existence of biofilm-related virulence genes of ompA, bap, blaPER-1, csgA. Therefore, it has become necessary to take the control measures to limit the development of A. baumannii.
Keywords: Acinetobacter baumannii; Antimicrobial resistance; Biofilm related-genes; Frequency; Peptide mass analysis; Raw meat.
© 2020 The Author(s).
Figures
References
-
- Ahmad A., Akhtar F., Sheikh A.A., Tipu M.Y., Nasar M. Comparative antibiotic resistance profile of Acinetobacter species, isolated from fish, chicken and beef meat. J. Clin. Microbiol. Antimicrob. 2018;2(1):1.
-
- Aliramezani A., Douraghi M., Hajihasani A., Mohammadzadeh M., Rahbar M. Clonal relatedness and biofilm formation of OXA-23-producing carbapenem resistant Acinetobacter baumannii isolates from hospital environment. Micobial. Pathog. 2016;99:204–208. - PubMed
-
- Amorim A.M., Nascimento J.D. Acinetobacter: an underrated foodborne pathogen? J. Infect. Dev. Ctries. 2017;11(2):111–114. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

