Single-Cell Transcriptomics Reveals That Metabolites Produced by Paenibacillus bovis sp. nov. BD3526 Ameliorate Type 2 Diabetes in GK Rats by Downregulating the Inflammatory Response
- PMID: 33424779
- PMCID: PMC7793688
- DOI: 10.3389/fmicb.2020.568805
Single-Cell Transcriptomics Reveals That Metabolites Produced by Paenibacillus bovis sp. nov. BD3526 Ameliorate Type 2 Diabetes in GK Rats by Downregulating the Inflammatory Response
Abstract
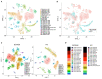
Chronic low-grade inflammation is widely involved in the development and progression of metabolic syndrome, which can lead to type 2 diabetes mellitus (T2DM). Dysregulation of proinflammatory and anti-inflammatory cytokines not only impairs insulin secretion by pancreatic β-cells but also results in systemic complications in late diabetes. In our previous work, metabolites produced by Paenibacillus bovis sp. nov. BD3526, which were isolated from Tibetan yak milk, demonstrated antidiabetic effects in Goto-Kakizaki (GK) rats. In this work, we used single-cell RNA sequencing (scRNA-seq) to further explore the impact of BD3526 metabolites on the intestinal cell composition of GK rats. Oral administration of the metabolites significantly reduced the number of adipocytes in the colon tissue of GK rats. In addition, cluster analysis of immune cells confirmed that the metabolites reduced the expression of interleukin (IL)-1β in macrophages in the colon and increased the numbers of dendritic cells (DCs) and regulatory T (Treg) cells. Further mechanistic studies of DCs confirmed that activation of the WNT/β-catenin pathway in DCs promoted the expression of IL-10 and transforming growth factor (TGF)-β, thereby increasing the number of Treg cells.
Keywords: Paenibacillus bovis sp. nov. BD3526; immune regulation; intestinal barrier; single-cell transcriptome sequencing; type 2 diabetes.
Copyright © 2020 Qiao, Wang, Zhang, Han, Feng and Wu.
Conflict of interest statement
ZQ, JH, XW, HZ, HF, and ZW are employed by Bright Dairy and Food Co.
Figures






References
-
- Boni-Schnetzler M., Jeffrey T., Geraldine P., Lorella M., Ehses J. A., Julie K. C., et al. (2008). Increased interleukin (IL)-1β messenger ribonucleic acid expression in β-cells of individuals with type 2 diabetes and regulation of IL-1β in human islets by glucose and autostimulation. J. Clin. Endocrinol. Metab. 93, 4065–4074. 10.1210/jc.2008-0396 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

