Characterization of Metagenome-Assembled Genomes and Carbohydrate-Degrading Genes in the Gut Microbiota of Tibetan Pig
- PMID: 33424798
- PMCID: PMC7785962
- DOI: 10.3389/fmicb.2020.595066
Characterization of Metagenome-Assembled Genomes and Carbohydrate-Degrading Genes in the Gut Microbiota of Tibetan Pig
Abstract
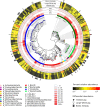
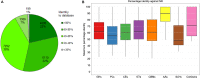
Tibetan pig is an important domestic mammal, providing products of high nutritional value for millions of people living in the Qinghai-Tibet Plateau. The genomes of mammalian gut microbiota encode a large number of carbohydrate-active enzymes, which are essential for the digestion of complex polysaccharides through fermentation. However, the current understanding of microbial degradation of dietary carbohydrates in the Tibetan pig gut is limited. In this study, we produced approximately 145 gigabases of metagenomic sequence data for the fecal samples from 11 Tibetan pigs. De novo assembly and binning recovered 322 metagenome-assembled genomes taxonomically assigned to 11 bacterial phyla and two archaeal phyla. Of these genomes, 191 represented the uncultivated microbes derived from novel prokaryotic taxa. Twenty-three genomes were identified as metagenomic biomarkers that were significantly abundant in the gut ecosystem of Tibetan pigs compared to the other low-altitude relatives. Further, over 13,000 carbohydrate-degrading genes were identified, and these genes were more abundant in some of the genomes within the five principal phyla: Firmicutes, Bacteroidetes, Spirochaetota, Verrucomicrobiota, and Fibrobacterota. Particularly, three genomes representing the uncultivated Verrucomicrobiota encode the most abundant degradative enzymes in the fecal microbiota of Tibetan pigs. These findings should substantially increase the phylogenetic diversity of specific taxonomic clades in the microbial tree of life and provide an expanded repertoire of biomass-degrading genes for future application to microbial production of industrial enzymes.
Keywords: Tibetan pig; carbohydrate-degrading genes; complex carbohydrates; gut microbiota; metagenome-assembled genomes; uncultivated microorganisms.
Copyright © 2020 Zhou, Luo, Gong, Wang, Gesang, Wang, Xu and Suolang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

