Physical Characterization of Colorectal Cancer Spheroids and Evaluation of NK Cell Infiltration Through a Flow-Based Analysis
- PMID: 33424829
- PMCID: PMC7786051
- DOI: 10.3389/fimmu.2020.564887
Physical Characterization of Colorectal Cancer Spheroids and Evaluation of NK Cell Infiltration Through a Flow-Based Analysis
Abstract
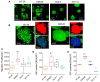
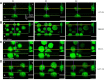
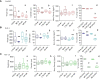
To improve pathogenetic studies in cancer development and reliable preclinical testing of anti-cancer treatments, three-dimensional (3D) cultures, including spheroids, have been widely recognized as more physiologically relevant in vitro models of in vivo tumor behavior. Currently, the generation of uniformly sized spheroids is still challenging: different 3D cell culture methods produce heterogeneous populations in dimensions and morphology, that may strongly influence readouts reliability correlated to tumor growth rate or antitumor natural killer (NK) cell-mediated cytotoxicity. In this context, an increasing consensus claims the integration of microfluidic technologies within 3D cell culture, as the physical characterization of tumor spheroids is unavoidably demanded to standardize protocols and assays for in vitro testing. In this paper, we employed a flow-based method specifically conceived to measure weight, size and focused onto mass density values of tumor spheroids. These measurements are combined with confocal and digital imaging of such samples. We tested the spheroids of four colorectal cancer (CRC) cell lines that exhibit statistically relevant differences in their physical characteristics, even though starting from the same cell seeding density. These variations are seemingly cell line-dependent and associated with the number of growing cells and the degree of spheroid compaction as well, supported by different adenosine-triphosphate contents. We also showed that this technology can estimate the NK cell killing efficacy by measuring the weight loss and diameter shrinkage of tumor spheroids, alongside with the commonly used cell viability in vitro test. As the activity of NK cells relies on their infiltration rate, the in vitro sensitivity of CRC spheroids proved to be exposure time- and cell line-dependent with direct correlation to the cell viability reduction. All these functional aspects can be measured by the system and are documented by digital image analysis. In conclusion, this flow-based method potentially paves the way towards standardization of 3D cell cultures and its early adoption in cancer research to test antitumor immune response and set up new immunotherapy strategies.
Keywords: 3D cell culture; colorectal cancer; mass density; microfluidics; natural killer cells; spheroid; weight.
Copyright © 2020 Sargenti, Musmeci, Bacchi, Delprete, Cristaldi, Cannas, Bonetti, Pasqua, Gazzola, Costa, Villa, Zocchi and Poggi.
Conflict of interest statement
The authors of Affiliation 1 (AS, FM, FB, DC, FC, SB, SP, and DG) are employed by Cell Dynamics isrl company. The authors declare that a Patent Application (No. 102020000006031) incorporating parts of this work has been filed. DG, SB, DC, AS, and FM are the inventors of patent No. 102020000006031. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Cocultures of human colorectal tumor spheroids with immune cells reveal the therapeutic potential of MICA/B and NKG2A targeting for cancer treatment.J Immunother Cancer. 2019 Mar 14;7(1):74. doi: 10.1186/s40425-019-0553-9. J Immunother Cancer. 2019. PMID: 30871626 Free PMC article.
-
Cytotoxicity and infiltration of human NK cells in in vivo-like tumor spheroids.BMC Cancer. 2015 May 3;15:351. doi: 10.1186/s12885-015-1321-y. BMC Cancer. 2015. PMID: 25933805 Free PMC article.
-
Zoledronate Triggers Vδ2 T Cells to Destroy and Kill Spheroids of Colon Carcinoma: Quantitative Image Analysis of Three-Dimensional Cultures.Front Immunol. 2018 May 8;9:998. doi: 10.3389/fimmu.2018.00998. eCollection 2018. Front Immunol. 2018. PMID: 29867975 Free PMC article.
-
Uniform sized cancer spheroids production using hydrogel-based droplet microfluidics: a review.Biomed Microdevices. 2024 May 29;26(2):26. doi: 10.1007/s10544-024-00712-3. Biomed Microdevices. 2024. PMID: 38806765 Free PMC article. Review.
-
Generation of Multicellular Breast Cancer Tumor Spheroids: Comparison of Different Protocols.J Mammary Gland Biol Neoplasia. 2016 Dec;21(3-4):89-98. doi: 10.1007/s10911-016-9359-2. Epub 2016 Aug 12. J Mammary Gland Biol Neoplasia. 2016. PMID: 27518775 Review.
Cited by
-
Adoptive NK Cell Transfer as a Treatment in Colorectal Cancer Patients: Analyses of Tumour Cell Determinants Correlating With Efficacy In Vitro and In Vivo.Front Immunol. 2022 Jun 7;13:890836. doi: 10.3389/fimmu.2022.890836. eCollection 2022. Front Immunol. 2022. PMID: 35747143 Free PMC article.
-
A Descriptive Study of the Physical Direct Interaction between Adipose Tissue-Mesenchymal Stem Cells and Colo 205 Cells: Impact on Cancer Cells Stemness, and Intracellular Reactive Oxygen Species Levels.Asian Pac J Cancer Prev. 2022 May 1;23(5):1635-1645. doi: 10.31557/APJCP.2022.23.5.1635. Asian Pac J Cancer Prev. 2022. PMID: 35633548 Free PMC article.
-
HAMP as a Prognostic Biomarker for Colorectal Cancer Based on Tumor Microenvironment Analysis.Front Oncol. 2022 Aug 5;12:884474. doi: 10.3389/fonc.2022.884474. eCollection 2022. Front Oncol. 2022. PMID: 35992796 Free PMC article.
-
Adipose Stromal Cell Spheroids for Cartilage Repair: A Promising Tool for Unveiling the Critical Maturation Point.Bioengineering (Basel). 2023 Oct 12;10(10):1182. doi: 10.3390/bioengineering10101182. Bioengineering (Basel). 2023. PMID: 37892912 Free PMC article.
-
Pediatric Tumors-Mediated Inhibitory Effect on NK Cells: The Case of Neuroblastoma and Wilms' Tumors.Cancers (Basel). 2021 May 14;13(10):2374. doi: 10.3390/cancers13102374. Cancers (Basel). 2021. PMID: 34069127 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

