Mitochondrial Fostering: The Mitochondrial Genome May Play a Role in Plant Orphan Gene Evolution
- PMID: 33424897
- PMCID: PMC7793901
- DOI: 10.3389/fpls.2020.600117
Mitochondrial Fostering: The Mitochondrial Genome May Play a Role in Plant Orphan Gene Evolution
Abstract
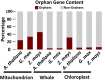
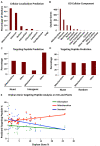
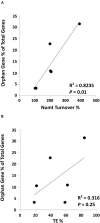
Plant mitochondrial genomes exhibit unique evolutionary patterns. They have a high rearrangement but low mutation rate, and a large size. Based on massive mitochondrial DNA transfers to the nucleus as well as the mitochondrial unique evolutionary traits, we propose a "Mitochondrial Fostering" theory where the organelle genome plays an integral role in the arrival and development of orphan genes (genes with no homologs in other lineages). Two approaches were used to test this theory: (1) bioinformatic analysis of nuclear mitochondrial DNA (Numts: mitochondrial originating DNA that migrated to the nucleus) at the genome level, and (2) bioinformatic analysis of particular orphan sequences present in both the mitochondrial genome and the nuclear genome of Arabidopsis thaliana. One study example is given about one orphan sequence that codes for two unique orphan genes: one in the mitochondrial genome and another one in the nuclear genome. DNA alignments show regions of this A. thaliana orphan sequence exist scattered throughout other land plant mitochondrial genomes. This is consistent with the high recombination rates of mitochondrial genomes in land plants. This may also enable the creation of novel coding sequences within the orphan loci, which can then be transferred to the nuclear genome and become exposed to new evolutionary pressures. Our study also reveals a high correlation between the amount of mitochondrial DNA transferred to the nuclear genome and the number of orphan genes in land plants. All the data suggests the mitochondrial genome may play a role in nuclear orphan gene evolution in land plants.
Keywords: evolution; mitochondrial genome; numt; orphan gene; plant.
Copyright © 2020 O’Conner and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

