Appropriate Activity Assays Are Crucial for the Specific Determination of Proline Dehydrogenase and Pyrroline-5-Carboxylate Reductase Activities
- PMID: 33424902
- PMCID: PMC7785524
- DOI: 10.3389/fpls.2020.602939
Appropriate Activity Assays Are Crucial for the Specific Determination of Proline Dehydrogenase and Pyrroline-5-Carboxylate Reductase Activities
Abstract
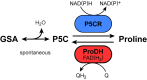
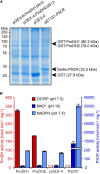
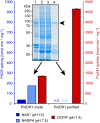
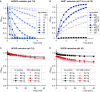
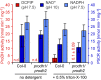
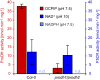
Accumulation of proline is a widespread plant response to a broad range of environmental stress conditions including salt and osmotic stress. Proline accumulation is achieved mainly by upregulation of proline biosynthesis in the cytosol and by inhibition of proline degradation in mitochondria. Changes in gene expression or activity levels of the two enzymes catalyzing the first reactions in these two pathways, namely pyrroline-5-carboxylate (P5C) synthetase and proline dehydrogenase (ProDH), are often used to assess the stress response of plants. The difficulty to isolate ProDH in active form has led several researchers to erroneously report proline-dependent NAD+ reduction at pH 10 as ProDH activity. We demonstrate that this activity is due to P5C reductase (P5CR), the second and last enzyme in proline biosynthesis, which works in the reverse direction at unphysiologically high pH. ProDH does not use NAD+ as electron acceptor but can be assayed with the artificial electron acceptor 2,6-dichlorophenolindophenol (DCPIP) after detergent-mediated solubilization or enrichment of mitochondria. Seemingly counter-intuitive results from previous publications can be explained in this way and our data highlight the importance of appropriate and specific assays for the detection of ProDH and P5CR activities in crude plant extracts.
Keywords: electron acceptor; enzyme activity assay; mitochondria; proline dehydrogenase; protein extraction; pyrroline-5-carboxylate reductase.
Copyright © 2020 Lebreton, Cabassa-Hourton, Savouré, Funck and Forlani.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Regulation of levels of proline as an osmolyte in plants under water stress.Plant Cell Physiol. 1997 Oct;38(10):1095-102. doi: 10.1093/oxfordjournals.pcp.a029093. Plant Cell Physiol. 1997. PMID: 9399433 Review.
-
Unraveling delta1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes.J Biol Chem. 2009 Sep 25;284(39):26482-92. doi: 10.1074/jbc.M109.009340. Epub 2009 Jul 27. J Biol Chem. 2009. PMID: 19635803 Free PMC article.
-
Proline dehydrogenase contributes to pathogen defense in Arabidopsis.Plant Physiol. 2011 Apr;155(4):1947-59. doi: 10.1104/pp.110.167163. Epub 2011 Feb 10. Plant Physiol. 2011. PMID: 21311034 Free PMC article.
-
Context of action of proline dehydrogenase (ProDH) in the Hypersensitive Response of Arabidopsis.BMC Plant Biol. 2014 Jan 13;14:21. doi: 10.1186/1471-2229-14-21. BMC Plant Biol. 2014. PMID: 24410747 Free PMC article.
-
Prolidase-proline dehydrogenase/proline oxidase-collagen biosynthesis axis as a potential interface of apoptosis/autophagy.Biofactors. 2016 Jul 8;42(4):341-8. doi: 10.1002/biof.1283. Epub 2016 Apr 4. Biofactors. 2016. PMID: 27040799 Review.
Cited by
-
The Proline Dehydrogenase Gene CsProDH1 Regulates Homeostasis of the Pro-P5C Cycle Under Drought Stress in Tea Plants.Int J Mol Sci. 2025 Mar 28;26(7):3121. doi: 10.3390/ijms26073121. Int J Mol Sci. 2025. PMID: 40243904 Free PMC article.
-
Kinetics of human pyrroline-5-carboxylate reductase in L-thioproline metabolism.Amino Acids. 2021 Dec;53(12):1863-1874. doi: 10.1007/s00726-021-03095-4. Epub 2021 Nov 18. Amino Acids. 2021. PMID: 34792644 Free PMC article.
-
Transcriptome analysis reveals the proline metabolic pathway and its potential regulation TF-hub genes in salt-stressed potato.Front Plant Sci. 2022 Oct 17;13:1030138. doi: 10.3389/fpls.2022.1030138. eCollection 2022. Front Plant Sci. 2022. PMID: 36325562 Free PMC article.
-
Interplay between Proline Metabolism and ROS in the Fine Tuning of Root-Meristem Size in Arabidopsis.Plants (Basel). 2022 Jun 5;11(11):1512. doi: 10.3390/plants11111512. Plants (Basel). 2022. PMID: 35684285 Free PMC article.
-
The nonphototrophic hypocotyl 3 (NPH3) domain protein NRL5 is a trafficking-associated GTPase essential for drought resistance.Sci Adv. 2024 Aug 9;10(32):eado5429. doi: 10.1126/sciadv.ado5429. Epub 2024 Aug 9. Sci Adv. 2024. PMID: 39121213 Free PMC article.
References
-
- Basford R. E., Huennekens F. M. (1955). Studies on Thiols. I. Oxidation of Thiol groups by 2, 6-Dichlorophenol Indophenol1. J. Am. Chem. Soc. 77 3873–3877. 10.1021/ja01619a058 - DOI
-
- Cabassa-Hourton C., Schertl P., Bordenave-Jacquemin M., Saadallah K., Guivarc’h A., Lebreton S., et al. (2016). Proteomic and functional analysis of proline dehydrogenase 1 link proline catabolism to mitochondrial electron transport in Arabidopsis thaliana. Biochem. J. 473 2623–2634. 10.1042/bcj20160314 - DOI - PubMed
LinkOut - more resources
Full Text Sources

