Clovamide, a Hydroxycinnamic Acid Amide, Is a Resistance Factor Against Phytophthora spp. in Theobroma cacao
- PMID: 33424909
- PMCID: PMC7786005
- DOI: 10.3389/fpls.2020.617520
Clovamide, a Hydroxycinnamic Acid Amide, Is a Resistance Factor Against Phytophthora spp. in Theobroma cacao
Abstract
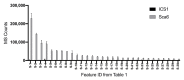
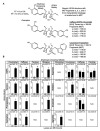
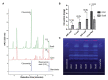
The hydroxycinnamic acid amides (HCAAs) are a diverse group of plant-specialized phenylpropanoid metabolites distributed widely in the plant kingdom and are known to be involved in tolerance to abiotic and biotic stress. The HCAA clovamide is reported in a small number of distantly related species. To explore the contribution of specialized metabolites to disease resistance in cacao (Theobroma cacao L., chocolate tree), we performed untargeted metabolomics using liquid chromatography - tandem mass spectrometry (LC-MS/MS) and compared the basal metabolite profiles in leaves of two cacao genotypes with contrasting levels of susceptibility to Phytophthora spp. Leaves of the tolerant genotype 'Scavina 6' ('Sca6') were found to accumulate dramatically higher levels of clovamide and several other HCAAs compared to the susceptible 'Imperial College Selection 1' ('ICS1'). Clovamide was the most abundant metabolite in 'Sca6' leaf extracts based on MS signal, and was up to 58-fold higher in 'Sca6' than in 'ICS1'. In vitro assays demonstrated that clovamide inhibits growth of three pathogens of cacao in the genus Phytophthora, is a substrate for cacao polyphenol oxidase, and is a contributor to enzymatic browning. Furthermore, clovamide inhibited proteinase and pectinase in vitro, activities associated with defense in plant-pathogen interactions. Fruit epidermal peels from both genotypes contained substantial amounts of clovamide, but two sulfated HCAAs were present at high abundance exclusively in 'Sca6' suggesting a potential functional role of these compounds. The potential to breed cacao with increased HCAAs for improved agricultural performance is discussed.
Keywords: Phytophthora; Theobroma cacao; black pod rot; clovamide; hydroxycinnamic acid amide; metabolomics; oomycete; polyphenol oxidase.
Copyright © 2020 Knollenberg, Li, Lambert, Maximova and Guiltinan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Akaza J. M., Kouassi A. B., Akaffou D. S., Fouet O., N’Guetta A. S. -P., Lanaud C. (2016). Mapping QTLs for black pod (Phytophthora palmivora) resistance in three hybrid progenies of cocoa (Theobroma cacao L.) using SSR markers. Int. J. Sci. Res. Publ. 6, 298–311.
-
- Akaza M. J., N’Goran J. A. K., N’Guetta S. P. A., Kébé B. I., Tahi G. M., Sangaré A. (2009). Resistance to Phytophthora palmivora (Butler) Butler assessed on leaf discs of cacao (Theobroma cacao L.) hybrid trees. Asian J. Plant Pathol. 3, 106–118. 10.3923/ajppaj.2009.106.118 - DOI
-
- Ali S. S., Shao J., Lary D. J., Kronmiller B. A., Shen D., Strem M. D., et al. (2017). Phytophthora megakarya and Phytophthora palmivora, closely related causal agents of cacao black pod rot, underwent increases in genome sizes and gene numbers by different mechanisms. Genome Biol. Evol. 9, 536–557. 10.1093/gbe/evx021, PMID: - DOI - PMC - PubMed
-
- Alonso-Salces R. M., Guillou C., Berrueta L. A. (2009). Liquid chromatography coupled with ultraviolet absorbance detection, electrospray ionization, collision-induced dissociation and tandem mass spectrometry on a triple quadrupole for the on-line characterization of polyphenols and methylxanthines in green coffee beans. Rapid Commun. Mass Spectrom. 23, 363–383. 10.1002/rcm.3884, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

