Genome-Wide Analysis of Cell-Free DNA Methylation Profiling for the Early Diagnosis of Pancreatic Cancer
- PMID: 33424927
- PMCID: PMC7794002
- DOI: 10.3389/fgene.2020.596078
Genome-Wide Analysis of Cell-Free DNA Methylation Profiling for the Early Diagnosis of Pancreatic Cancer
Abstract
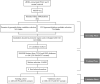
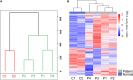
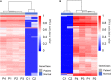
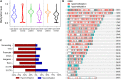
As one of the most malicious cancers, pancreatic cancer is difficult to treat due to the lack of effective early diagnosis. Therefore, it is urgent to find reliable diagnostic and predictive markers for the early detection of pancreatic cancer. In recent years, the detection of circulating cell-free DNA (cfDNA) methylation in plasma has attracted global attention for non-invasive and early cancer diagnosis. Here, we carried out a genome-wide cfDNA methylation profiling study of pancreatic ductal adenocarcinoma (PDAC) patients by methylated DNA immunoprecipitation coupled with high-throughput sequencing (MeDIP-seq). Compared with healthy individuals, 775 differentially methylated regions (DMRs) located in promoter regions were identified in PDAC patients with 761 hypermethylated and 14 hypomethylated regions; meanwhile, 761 DMRs in CpG islands (CGIs) were identified in PDAC patients with 734 hypermethylated and 27 hypomethylated regions (p-value < 0.0001). Then, 143 hypermethylated DMRs were further selected which were located in promoter regions and completely overlapped with CGIs. After performing the least absolute shrinkage and selection operator (LASSO) method, a total of eight markers were found to fairly distinguish PDAC patients from healthy individuals, including TRIM73, FAM150A, EPB41L3, SIX3, MIR663, MAPT, LOC100128977, and LOC100130148. In conclusion, this work identified a set of eight differentially methylated markers that may be potentially applied in non-invasive diagnosis of pancreatic cancer.
Keywords: MeDIP-seq; biomarkers; cfDNA; methylation; pancreatic ductal adenocarcinoma.
Copyright © 2020 Li, Wang, Zhao, Wang, Lu, Kang, Jin and Tian.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures



Similar articles
-
Genome-wide analysis of cell-Free DNA methylation profiling with MeDIP-seq identified potential biomarkers for colorectal cancer.World J Surg Oncol. 2022 Jan 22;20(1):21. doi: 10.1186/s12957-022-02487-4. World J Surg Oncol. 2022. PMID: 35065650 Free PMC article.
-
Genome-Wide Plasma Cell-Free DNA Methylation Profiling Identifies Potential Biomarkers for Lung Cancer.Dis Markers. 2019 Feb 5;2019:4108474. doi: 10.1155/2019/4108474. eCollection 2019. Dis Markers. 2019. PMID: 30867848 Free PMC article.
-
High-frequency aberrantly methylated targets in pancreatic adenocarcinoma identified via global DNA methylation analysis using methylCap-seq.Clin Epigenetics. 2014 Sep 22;6(1):18. doi: 10.1186/1868-7083-6-18. eCollection 2014. Clin Epigenetics. 2014. PMID: 25276247 Free PMC article.
-
Computational challenges in detection of cancer using cell-free DNA methylation.Comput Struct Biotechnol J. 2021 Dec 7;20:26-39. doi: 10.1016/j.csbj.2021.12.001. eCollection 2022. Comput Struct Biotechnol J. 2021. PMID: 34976309 Free PMC article. Review.
-
Epigenetic Landscape of DNA Methylation in Pancreatic Ductal Adenocarcinoma.Epigenomes. 2024 Nov 3;8(4):41. doi: 10.3390/epigenomes8040041. Epigenomes. 2024. PMID: 39584964 Free PMC article. Review.
Cited by
-
Advancements in Early Detection and Screening Strategies for Pancreatic Cancer: From Genetic Susceptibility to Novel Biomarkers.J Clin Med. 2024 Aug 10;13(16):4706. doi: 10.3390/jcm13164706. J Clin Med. 2024. PMID: 39200847 Free PMC article. Review.
-
Probabilistic modeling methods for cell-free DNA methylation based cancer classification.BMC Bioinformatics. 2022 Apr 4;23(1):119. doi: 10.1186/s12859-022-04651-9. BMC Bioinformatics. 2022. PMID: 35379172 Free PMC article.
-
Diagnosing and monitoring pancreatic cancer through cell-free DNA methylation: progress and prospects.Biomark Res. 2023 Oct 5;11(1):88. doi: 10.1186/s40364-023-00528-y. Biomark Res. 2023. PMID: 37798621 Free PMC article. Review.
-
DNA methylation analysis explores the molecular basis of plasma cell-free DNA fragmentation.Nat Commun. 2023 Jan 18;14(1):287. doi: 10.1038/s41467-023-35959-6. Nat Commun. 2023. PMID: 36653380 Free PMC article.
-
Diagnostic methods for pancreatic cancer and their clinical applications (Review).Oncol Lett. 2025 May 27;30(1):370. doi: 10.3892/ol.2025.15116. eCollection 2025 Jul. Oncol Lett. 2025. PMID: 40496545 Free PMC article. Review.
References
-
- Benjamini Y., Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29 1165–1188.
-
- Chu L. C., Goggins M. G., Fishman E. K. (2017). Diagnosis and detection of pancreatic cancer. Cancer J. 23 333–342. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

