Genetic Determinism Exists for the Global DNA Methylation Rate in Sheep
- PMID: 33424937
- PMCID: PMC7786236
- DOI: 10.3389/fgene.2020.616960
Genetic Determinism Exists for the Global DNA Methylation Rate in Sheep
Abstract
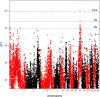
Recent studies showed that epigenetic marks, including DNA methylation, influence production and adaptive traits in plants and animals. So far, most studies dealing with genetics and epigenetics considered DNA methylation sites independently. However, the genetic basis of the global DNA methylation rate (GDMR) remains unknown. The main objective of the present study was to investigate genetic determinism of GDMR in sheep. The experiment was conducted on 1,047 Romane sheep allocated into 10 half-sib families. After weaning, all the lambs were phenotyped for global GDMR in blood as well as for production and adaptive traits. GDMR was measured by LUminometric Methylation Analysis (LUMA) using a pyrosequencing approach. Association analyses were conducted on some of the lambs (n = 775) genotyped by using the Illumina OvineSNP50 BeadChip. Blood GDMR varied among the animals (average 70.7 ± 6.0%). Female lambs had significantly higher GDMR than male lambs. Inter-individual variability of blood GDMR had an additive genetic component and heritability was moderate (h 2 = 0.20 ± 0.05). No significant genetic correlation was found between GDMR and growth or carcass traits, birthcoat, or social behaviors. Association analyses revealed 28 QTLs associated with blood GDMR. Seven genomic regions on chromosomes 1, 5, 11, 17, 24, and 26 were of most interest due to either high significant associations with GDMR or to the relevance of genes located close to the QTLs. QTL effects were moderate. Genomic regions associated with GDMR harbored several genes not yet described as being involved in DNA methylation, but some are already known to play an active role in gene expression. In addition, some candidate genes, CHD1, NCO3A, KDM8, KAT7, and KAT6A have previously been described to be involved in epigenetic modifications. In conclusion, the results of the present study indicate that blood GDMR in domestic sheep is under polygenic influence and provide new insights into DNA methylation genetic determinism.
Keywords: QTL; blood; epigenetics; genetic correlation; heritability.
Copyright © 2020 Hazard, Plisson-Petit, Moreno-Romieux, Fabre and Drouilhet.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Variability in Global DNA Methylation Rate Across Tissues and Over Time in Sheep.Front Genet. 2022 Mar 11;13:791283. doi: 10.3389/fgene.2022.791283. eCollection 2022. Front Genet. 2022. PMID: 35360841 Free PMC article.
-
Identification of QTLs for behavioral reactivity to social separation and humans in sheep using the OvineSNP50 BeadChip.BMC Genomics. 2014 Sep 9;15(1):778. doi: 10.1186/1471-2164-15-778. BMC Genomics. 2014. PMID: 25204347 Free PMC article.
-
Importance of birthcoat for lamb survival and growth in the Romane sheep breed extensively managed on rangelands.J Anim Sci. 2014 Jan;92(1):54-63. doi: 10.2527/jas.2013-6660. Epub 2013 Dec 23. J Anim Sci. 2014. PMID: 24366070
-
[Epigenetics of schizophrenia: a review].Encephale. 2014 Oct;40(5):380-6. doi: 10.1016/j.encep.2014.06.005. Epub 2014 Aug 12. Encephale. 2014. PMID: 25127897 Review. French.
-
Epigenetics and arterial hypertension: the challenge of emerging evidence.Transl Res. 2015 Jan;165(1):154-65. doi: 10.1016/j.trsl.2014.06.007. Epub 2014 Jun 25. Transl Res. 2015. PMID: 25035152 Review.
Cited by
-
Beyond the genome: a perspective on the use of DNA methylation profiles as a tool for the livestock industry.Anim Front. 2021 Dec 17;11(6):90-94. doi: 10.1093/af/vfab060. eCollection 2021 Dec. Anim Front. 2021. PMID: 34934534 Free PMC article. Review. No abstract available.
-
Genetic regulation of sperm DNA methylation in cattle through meQTL mapping.BMC Genomics. 2025 Aug 22;26(1):771. doi: 10.1186/s12864-025-11934-x. BMC Genomics. 2025. PMID: 40847283 Free PMC article.
-
Variability in Global DNA Methylation Rate Across Tissues and Over Time in Sheep.Front Genet. 2022 Mar 11;13:791283. doi: 10.3389/fgene.2022.791283. eCollection 2022. Front Genet. 2022. PMID: 35360841 Free PMC article.
-
Editorial: Epigenetic Variation Influences on Livestock Production and Disease Traits.Front Genet. 2022 Jun 15;13:942747. doi: 10.3389/fgene.2022.942747. eCollection 2022. Front Genet. 2022. PMID: 35783264 Free PMC article. No abstract available.
-
Whole-genome DNA methylation profiling reveals epigenetic signatures in developing muscle in Tan and Hu sheep and their offspring.Front Vet Sci. 2023 Jun 14;10:1186040. doi: 10.3389/fvets.2023.1186040. eCollection 2023. Front Vet Sci. 2023. PMID: 37388464 Free PMC article.
References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate - a pratical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
LinkOut - more resources
Full Text Sources

