LncRNA DHRS4-AS1 Inhibits the Stemness of NSCLC Cells by Sponging miR-224-3p and Upregulating TP53 and TET1
- PMID: 33425890
- PMCID: PMC7786137
- DOI: 10.3389/fcell.2020.585251
LncRNA DHRS4-AS1 Inhibits the Stemness of NSCLC Cells by Sponging miR-224-3p and Upregulating TP53 and TET1
Abstract
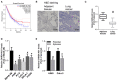
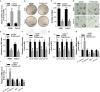
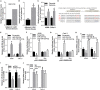
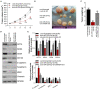
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related death. This study aimed to examine the roles of DHRS4-AS1/miR-224-3p signaling in the cancer cell stemness of NSCLC. Real-time PCR showed that DHRS4-AS1 was downregulated in cancerous tissues, and bioinformatics analysis revealed that high DHRS4-AS1 expression indicated a good prognosis for NSCLC patients. Sphere and colony formation assays showed that DHRS4-AS1 overexpression significantly suppressed NSCLC cell colony formation and stem cell-like properties. DHRS4-AS1 also abrogated the expression of OCT4, SOX2, CD34, and CD133, markedly inhibited the expression of epithelial-mesenchymal transition (EMT)-related factors, N-cadherin, ZEB1, and Vimentin, and increased E-cadherin expression in spheres. Furthermore, luciferase reporter assays and real-time PCR analysis demonstrated that DHRS4-AS1 and miR-224-3p were antagonistically repressed in NSCLC cells. RNA immunoprecipitation (RIP) analysis revealed that DHRS4-AS1 interacted with miR-224-3p. DHRS4-AS1 partially reversed the miR-224-3p-decreased TP53 and TET1, resulting in the inhibition of tumor growth in vivo. Finally, TP53 and TET1 were antagonistically regulated by DHRS4-AS1 and miR-224-3p in NSCLC cells. In conclusion, TP53- and TET1-associated DHRS4-AS1/miR-224-3p axis is an essential mechanism by which NSCLC modulates cancer cell stemness.
Keywords: DHRS4-AS1; TET1; TP53; cancer cell stemness; miR-224-3p; non-small cell lung cancer.
Copyright © 2020 Yan, Zhao, Xu, Li, Li, Liu, Shi and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
LncRNA ASAP1-IT1 enhances cancer cell stemness via regulating miR-509-3p/YAP1 axis in NSCLC.Cancer Cell Int. 2021 Oct 29;21(1):572. doi: 10.1186/s12935-021-02270-7. Cancer Cell Int. 2021. PMID: 34715859 Free PMC article.
-
Feasibility study of lncRNA DHRS4-AS1 sponge miR-222-3p in the diagnosis of thyroid cancer.Endokrynol Pol. 2024;75(5):494-500. doi: 10.5603/ep.99456. Epub 2024 Oct 8. Endokrynol Pol. 2024. PMID: 39376175
-
Highly expressed lncRNA FOXD3-AS1 promotes non-small cell lung cancer progression via regulating miR-127-3p/mediator complex subunit 28 axis.Eur Rev Med Pharmacol Sci. 2020 Mar;24(5):2525-2538. doi: 10.26355/eurrev_202003_20520. Eur Rev Med Pharmacol Sci. 2020. PMID: 32196603
-
LncRNA B4GALT1-AS1 promotes non-small cell lung cancer cell growth via increasing ZEB1 level by sponging miR-144-3p.Transl Cancer Res. 2022 Mar;11(3):538-547. doi: 10.21037/tcr-22-296. Transl Cancer Res. 2022. PMID: 35402178 Free PMC article.
-
microRNA-301b-3p downregulation underlies a novel inhibitory role of long non-coding RNA MBNL1-AS1 in non-small cell lung cancer.Stem Cell Res Ther. 2019 May 21;10(1):144. doi: 10.1186/s13287-019-1235-8. Stem Cell Res Ther. 2019. PMID: 31113460 Free PMC article.
Cited by
-
Long noncoding RNA lnc-SNAPC5-3:4 inhibits malignancy by directly upregulating miR-224-3p in non-small cell lung cancer.Heliyon. 2024 Jan 13;10(2):e24668. doi: 10.1016/j.heliyon.2024.e24668. eCollection 2024 Jan 30. Heliyon. 2024. PMID: 38312596 Free PMC article.
-
Triptolide reverses cis‑diamminedichloroplatinum resistance in esophageal squamous cell carcinoma by suppressing glycolysis and causing mitochondrial malfunction.Mol Med Rep. 2025 Mar;31(3):74. doi: 10.3892/mmr.2025.13439. Epub 2025 Jan 31. Mol Med Rep. 2025. PMID: 39886972 Free PMC article.
-
Microarray data analysis to identify miRNA biomarkers and construct the lncRNA-miRNA-mRNA network in lung adenocarcinoma.Medicine (Baltimore). 2022 Sep 9;101(36):e30393. doi: 10.1097/MD.0000000000030393. Medicine (Baltimore). 2022. PMID: 36086747 Free PMC article.
-
Screening microRNAs as potential prognostic biomarkers for lung adenocarcinoma.Ann Med. 2023;55(2):2241013. doi: 10.1080/07853890.2023.2241013. Epub 2023 Nov 6. Ann Med. 2023. PMID: 37930873 Free PMC article.
-
Identification and Roles of miR-29b-1-3p and miR29a-3p-Regulated and Non-Regulated lncRNAs in Endocrine-Sensitive and Resistant Breast Cancer Cells.Cancers (Basel). 2021 Jul 14;13(14):3530. doi: 10.3390/cancers13143530. Cancers (Basel). 2021. PMID: 34298743 Free PMC article.
References
-
- Bautista R. R., Gomez A. O., Miranda A. H., Dehesa A. Z., Villarreal-Garza C., Avila-Moreno F., et al. (2018). Correction to: long non-coding RNAs: implications in targeted diagnoses, prognosis, and improved therapeutic strategies in human non- and triple-negative breast cancer. Clin. Epigenetics 10:106 10.1186/s13148-018-0537-5 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

