Activation of a Mitogen-Activated Protein Kinase Hog1 by DNA Damaging Agent Methyl Methanesulfonate in Yeast
- PMID: 33425986
- PMCID: PMC7793754
- DOI: 10.3389/fmolb.2020.581095
Activation of a Mitogen-Activated Protein Kinase Hog1 by DNA Damaging Agent Methyl Methanesulfonate in Yeast
Abstract
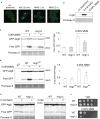
Hog1 is a mitogen-activated protein kinase in yeast that primarily regulates cellular responses to hyperosmolarity stress. In this study, we have examined the potential involvement of Hog1 in mediating cellular responses to DNA damaging agents. We find that treatment of yeast cells with DNA damaging agent methyl methanesulfonate (MMS) induces a marked and prolonged Hog1 activation. Distinct from stressors such as arsenite that activates Hog1 via inhibiting its phosphatases, activation of Hog1 by MMS is phosphatase-independent. Instead, MMS impairs a critical phosphor-relay process that normally keeps Hog1 in an inactive state. Functionally, MMS-activated Hog1 is not translocated to the nucleus to regulate gene expression but rather stays in the cytoplasm and regulates MMS-induced autophagy and cell adaptation to MMS stress. These findings reveal a new role of Hog1 in regulating MMS-induced cellular stress.
Keywords: DNA damage; Hog1; Ypd1; autophagy; mitogen-activated protein kinase; yeast Hog1 activation by DNA damage.
Copyright © 2020 Huang, Zhang, Weng and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Intracellular mechanism by which arsenite activates the yeast stress MAPK Hog1.Mol Biol Cell. 2018 Aug 1;29(15):1904-1915. doi: 10.1091/mbc.E18-03-0185. Epub 2018 May 30. Mol Biol Cell. 2018. PMID: 29846136 Free PMC article.
-
Differential activation of c-Jun NH2-terminal kinase and p38 mitogen-activated protein kinases by methyl methanesulfonate in the liver and brain of rats: implication for organ-specific carcinogenesis.Cancer Res. 2000 Sep 15;60(18):5067-73. Cancer Res. 2000. PMID: 11016630
-
Delayed Turnover of Unphosphorylated Ssk1 during Carbon Stress Activates the Yeast Hog1 Map Kinase Pathway.PLoS One. 2015 Sep 4;10(9):e0137199. doi: 10.1371/journal.pone.0137199. eCollection 2015. PLoS One. 2015. PMID: 26340004 Free PMC article.
-
Hog1: 20 years of discovery and impact.Sci Signal. 2014 Sep 16;7(343):re7. doi: 10.1126/scisignal.2005458. Sci Signal. 2014. PMID: 25227612 Review.
-
Stressing out or stressing in: intracellular pathways for SAPK activation.Curr Genet. 2019 Apr;65(2):417-421. doi: 10.1007/s00294-018-0898-5. Epub 2018 Oct 30. Curr Genet. 2019. PMID: 30377756 Free PMC article. Review.
Cited by
-
Hog1-mediated stress tolerance in the pathogenic fungus Trichosporon asahii.Sci Rep. 2023 Aug 19;13(1):13539. doi: 10.1038/s41598-023-40825-y. Sci Rep. 2023. PMID: 37598230 Free PMC article.
-
Hog1 acts in a Mec1-independent manner to counteract oxidative stress following telomerase inactivation in Saccharomyces cerevisiae.Commun Biol. 2024 Jun 22;7(1):761. doi: 10.1038/s42003-024-06464-3. Commun Biol. 2024. PMID: 38909140 Free PMC article.
-
Acetic acid-induced stress granules function as scaffolding complexes for Hog1 activation by Pbs2.J Cell Biol. 2025 May 5;224(5):e202409072. doi: 10.1083/jcb.202409072. Epub 2025 Mar 11. J Cell Biol. 2025. PMID: 40067148
-
MAP Kinase FgHog1 and Importin β FgNmd5 Regulate Calcium Homeostasis in Fusarium graminearum.J Fungi (Basel). 2023 Jun 28;9(7):707. doi: 10.3390/jof9070707. J Fungi (Basel). 2023. PMID: 37504696 Free PMC article.
-
Ca2+-triggered Atg11-Bmh1/2-Snf1 complex assembly initiates autophagy upon glucose starvation.J Cell Biol. 2024 Sep 2;223(9):e202310049. doi: 10.1083/jcb.202310049. Epub 2024 Jul 9. J Cell Biol. 2024. PMID: 38980288 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

