Exploring rotavirus proteome to identify potential B- and T-cell epitope using computational immunoinformatics
- PMID: 33426322
- PMCID: PMC7779714
- DOI: 10.1016/j.heliyon.2020.e05760
Exploring rotavirus proteome to identify potential B- and T-cell epitope using computational immunoinformatics
Abstract
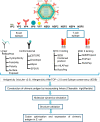
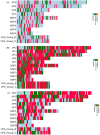
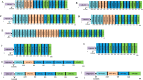
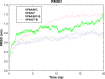
Rotavirus is the most common cause of acute gastroenteritis in infants and children worldwide. The functional correlation of B- and T-cells to long-lasting immunity against rotavirus infection in the literature is limited. In this work, a series of computational immuno-informatics approaches were applied and identified 28 linear B-cells, 26 conformational B-cell, 44 TC cell and 40 TH cell binding epitopes for structural and non-structural proteins of rotavirus. Further selection of putative B and T cell epitopes in the multi-epitope vaccine construct was carried out based on immunogenicity, conservancy, allergenicity and the helical content of predicted epitopes. An in-silico vaccine constructs was developed using an N-terminal adjuvant (RGD motif) followed by TC and TH cell epitopes and B-cell epitope with an appropriate linker. Multi-threading models of multi-epitope vaccine construct with B- and T-cell epitopes were generated and molecular dynamics simulation was performed to determine the stability of designed vaccine. Codon optimized multi-epitope vaccine antigens was expressed and affinity purified using the E. coli expression system. Further the T cell epitope presentation assay using the recombinant multi-epitope constructs and the T cell epitope predicted and identified in this study have not been investigated. Multi-epitope vaccine construct encompassing predicted B- and T-cell epitopes may help to generate long-term immune responses against rotavirus. The computational findings reported in this study may provide information in developing epitope-based vaccine and diagnostic assay for rotavirus-led diarrhea in children's.
Keywords: Immune epitope; Non-structural proteins; Rotavirus; Structural proteins.
© 2020 The Authors.
Figures






References
-
- Kapikian A.Z., Chanock R.M. Rotaviruses. In: Straus S.E., editor. third ed. Volume 2. Lippincott-Raven; Philadelphia: 1996. pp. 1657–1708. (Fields Virology).
-
- Butz A.M., Fosarelli P., Kick J., Yolken R. Prevalence of rotavirus on high-risk fomites in daycare facilities. Pediatrics. 1993;92(2):202–205. - PubMed
-
- Pesavento J.B., Crawford S.E., Estes M.K., Prasad B.V. Rotavirus proteins: structure and assembly. Curr. Top. Microbiol. Immunol. 2006;309:189–219. - PubMed
-
- Fields B.N., Knipe D.M., Howley P.M., Griffin D.E. Vol. 2. 2013. Fields virology; pp. 1347–1401. (Wolters Kluwer Health/Lippincott Williams & Wilkins). Chapter 45.
LinkOut - more resources
Full Text Sources
Other Literature Sources

