Methods for isolation and transcriptional profiling of individual cells from the human heart
- PMID: 33426328
- PMCID: PMC7779736
- DOI: 10.1016/j.heliyon.2020.e05810
Methods for isolation and transcriptional profiling of individual cells from the human heart
Abstract
Background: Global transcriptional profiling of individual cells represents a powerful approach to systematically survey contributions from cell-specific molecular phenotypes to human disease states but requires tissue-specific protocols. Here we sought to comprehensively evaluate protocols for single cell isolation and transcriptional profiling from heart tissue, focusing particularly on frozen tissue which is necessary for study of human hearts at scale.
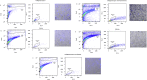
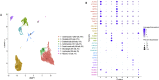
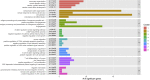
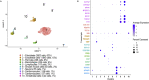
Methods and results: Using flow cytometry and high-content screening, we found that enzymatic dissociation of fresh murine heart tissue resulted in a sufficient yield of intact cells while for frozen murine or human heart resulted in low-quality cell suspensions across a range of protocols. These findings were consistent across enzymatic digestion protocols and whether samples were snap-frozen or treated with RNA-stabilizing agents before freezing. In contrast, we show that isolation of cardiac nuclei from frozen hearts results in a high yield of intact nuclei, and leverage expression arrays to show that nuclear transcriptomes reliably represent the cytoplasmic and whole-cell transcriptomes of the major cardiac cell types. Furthermore, coupling of nuclear isolation to PCM1-gated flow cytometry facilitated specific cardiomyocyte depletion, expanding resolution of the cardiac transcriptome beyond bulk tissue transcriptomes which were most strongly correlated with PCM1+ transcriptomes (r = 0.8). We applied these methods to generate a transcriptional catalogue of human cardiac cells by droplet-based RNA-sequencing of 8,460 nuclei from which cellular identities were inferred. Reproducibility of identified clusters was confirmed in an independent biopsy (4,760 additional PCM1- nuclei) from the same human heart.
Conclusion: Our results confirm the validity of single-nucleus but not single-cell isolation for transcriptional profiling of individual cells from frozen heart tissue, and establishes PCM1-gating as an efficient tool for cardiomyocyte depletion. In addition, our results provide a perspective of cell types inferred from single-nucleus transcriptomes that are present in an adult human heart.
Keywords: Cardiology; Cardiovascular System; Health sciences; Heart; Human; Methods; Protocol; Single cell; Transcriptome; Transcriptomics.
© 2020 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. - PMC - PubMed
-
- Asp M., Giacomello S., Larsson L., Wu C., Furth D., Qian X., Wardell E., Custodio J., Reimegard J., Salmen F., Osterholm C., Stahl P.L., Sundstrom E., Akesson E., Bergmann O., Bienko M., Mansson-Broberg A., Nilsson M., Sylven C., Lundeberg J. A spatiotemporal organ-wide gene expression and cell Atlas of the developing human heart. Cell. 2019;179:1647–1660. e19. - PubMed
-
- Bajpai G., Schneider C., Wong N., Bredemeyer A., Hulsmans M., Nahrendorf M., Epelman S., Kreisel D., Liu Y., Itoh A., Shankar T.S., Selzman C.H., Drakos S.G., Lavine K.J. The human heart contains distinct macrophage subsets with divergent origins and functions. Nat. Med. 2018;24:1234–1245. - PMC - PubMed
-
- Bakken T.E., Hodge R.D., Miller J.A., Yao Z., Nguyen T.N., Aevermann B., Barkan E., Bertagnolli D., Casper T., Dee N., Garren E., Goldy J., Graybuck L.T., Kroll M., Lasken R.S., Lathia K., Parry S., Rimorin C., Scheuermann R.H., Schork N.J. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PloS One. 2018;13 - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

