Metabolomic Signature of Human Aortic Valve Stenosis
- PMID: 33426374
- PMCID: PMC7775961
- DOI: 10.1016/j.jacbts.2020.10.001
Metabolomic Signature of Human Aortic Valve Stenosis
Abstract
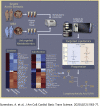
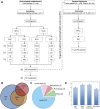
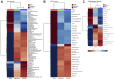
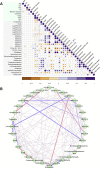
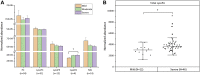
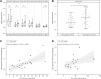
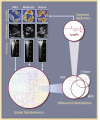
This study outlines the first step toward creating the metabolite atlas of human calcified aortic valves by identifying the expression of metabolites and metabolic pathways involved at various stages of calcific aortic valve stenosis progression. Untargeted analysis identified 72 metabolites and lipids that were significantly altered (p < 0.01) across different stages of disease progression. Of these metabolites and lipids, the levels of lysophosphatidic acid were shown to correlate with faster hemodynamic progression and could select patients at risk for faster progression rate.
Keywords: AS, aortic stenosis; ATX, autotaxin; AV, aortic valve; AVA, aortic valve area; BAV, bicuspid aortic valve; CAVS, calcific aortic valve stenosis; CV, correlation of variation; Lp(a), lipoprotein(a); LysoPA, lysophosphatidic acid; LysoPC, lysophosphatidylcholine; LysoPE, lysophosphatidylethanolamine; MG, monoglyceride; MPG, mean pressure gradient; PC, phosphatidylcholine; QC, quality control; TAV, tricuspid aortic valve; Vmax, peak aortic jet velocity; aortic stenosis; calcific aortic valve stenosis; lysophosphatidic acids; nontargeted metabolomics; targeted lipidomics; valvular calcification.
© 2020 The Authors.
Conflict of interest statement
Dr. Ravandi is supported by a grant from Research Manitoba and Heart and Stroke Foundation of Canada. Mr. Surendran is supported by Research Manitoba Master’s Studentship (2018), Bank of Montreal/Institute of Cardiovascular Sciences Studentship (2019), and Singal, Pawan K. Graduate Scholarship in Cardiovascular Sciences (2019). All other have reported that they have no relationships relevant to the contents of this paper to disclose.
Figures


Comment in
-
Calcific Aortic Valve Disease "Omics" Is Timely, But Are We Looking Too Late?JACC Basic Transl Sci. 2020 Dec 28;5(12):1178-1180. doi: 10.1016/j.jacbts.2020.11.001. eCollection 2020 Dec. JACC Basic Transl Sci. 2020. PMID: 33427821 Free PMC article.
References
-
- Nkomo V.T., Gardin J.M., Skelton T.N., Gottdiener J.S., Scott C.G., Enriquez-Sarano M. Burden of valvular heart diseases: a population-based study. Lancet. 2006;368:1005–1011. - PubMed
-
- Baumgartner H., Falk V., Bax J.J., ESC Scientific Document Group 2017 ESC/EACTS Guidelines for the management of valvular heart disease. Eur Heart J. 2017;38:2739–2791. - PubMed
-
- Arsenault B.J., Boekholdt S.M., Dubé M.P. Lipoprotein(a) levels, genotype, and incident aortic valve stenosis: a prospective Mendelian randomization study and replication in a case-control cohort. Circ Cardiovasc Genet. 2014;7:304–310. - PubMed
-
- Kamstrup P.R., Tybjaerg-Hansen A., Nordestgaard B.G. Elevated lipoprotein(a) and risk of aortic valve stenosis in the general population. J Am Coll Cardiol. 2014;63:470–477. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

