A multi-pronged approach targeting SARS-CoV-2 proteins using ultra-large virtual screening
- PMID: 33426509
- PMCID: PMC7783459
- DOI: 10.1016/j.isci.2020.102021
A multi-pronged approach targeting SARS-CoV-2 proteins using ultra-large virtual screening
Abstract
The unparalleled global effort to combat the continuing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic over the last year has resulted in promising prophylactic measures. However, a need still exists for cheap, effective therapeutics, and targeting multiple points in the viral life cycle could help tackle the current, as well as future, coronaviruses. Here, we leverage our recently developed, ultra-large-scale in silico screening platform, VirtualFlow, to search for inhibitors that target SARS-CoV-2. In this unprecedented structure-based virtual campaign, we screened roughly 1 billion molecules against each of 40 different target sites on 17 different potential viral and host targets. In addition to targeting the active sites of viral enzymes, we also targeted critical auxiliary sites such as functionally important protein-protein interactions.
Keywords: Drugs; High-Performance Computing in Bioinformatics; Structural Biology; Virology.
© 2020 The Author(s).
Conflict of interest statement
M.C. and V.D. report working for Innophore. C.C.G. reports being a shareholder and CEO of Innophore, an enzyme and drug discovery company. G.W. and C.G. report being co-founders of the company Virtual Discovery, Inc., which provides virtual screening services. G.W. reports serving as the director of this company. G.W. reports being a co-founder of PIC Therapeutics, Cellmig Biolabs, and Skinap Therapeutics. H.A. reports being an equity holder in PIC Therapeutics. I.I. and D.R. report working for Enamine, a company that is involved in the synthesis and distribution of chemical building blocks, fragments, and screening compounds. Y.M. reports being a scientific advisor for Enamine. Y.M., O.T., and A.P. report working for Chemspace, a company that is involved in the distribution of chemical building blocks, fragments, and screening compounds. I.D. reports working for UkrOrgSyntez Ltd. (UORSY), a company that is involved in the synthesis of chemical building blocks, fragments, and screening compounds. G.T., S.P., A.S., M.G., N.L., C.H., E.Y., R.L., R.Y., D.P., and J.K. report working for Google, a company that also provides cloud computing services. The research described here is scientifically and financially independent of the efforts in any of the above mentioned companies.
Figures
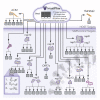



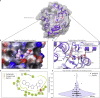
Update of
-
A Multi-Pronged Approach Targeting SARS-CoV-2 Proteins Using Ultra-Large Virtual Screening.ChemRxiv [Preprint]. 2020 Jul 24. doi: 10.26434/chemrxiv.12682316. ChemRxiv. 2020. Update in: iScience. 2021 Feb 19;24(2):102021. doi: 10.1016/j.isci.2020.102021. PMID: 33200116 Free PMC article. Updated. Preprint.
References
-
- Afar D.E., Vivanco I., Hubert R.S., Kuo J., Chen E., Saffran D.C., Raitano A.B., Jakobovits A. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001;61:1686–1692. - PubMed
-
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R. Coronavirus susceptibility to the antiviral Remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. - DOI - PMC - PubMed
-
- Agostini M.L., Pruijssers A.J., Chappell J.D., Gribble J., Lu X., Erica L.A., Bluemling G.R., Lockwood M.A., Sheahan T.P., Sims A.C., Natchus M.G. Small-molecule antiviral β-d-n4-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 2019;93 doi: 10.1128/jvi.01348-19. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

